Abstract
Early diagnosis plays a key role in prevention and treatment of skin cancer. Several machine learning techniques for accurate detection of skin cancer from medical images have been reported. Many of these techniques are based on pre-trained convolutional neural networks (CNNs), which enable training the models based on limited amounts of training data. However, the classification accuracy of these models still tends to be severely limited by the scarcity of representative images from malignant tumours. We propose a novel ensemble-based convolutional neural network (CNN) architecture where multiple CNN models, some of which are pre-trained and some are trained only on the data at hand, along with auxiliary data in the form of metadata associated with the input images, are combined using a meta-learner. The proposed approach improves the model’s ability to handle limited and imbalanced data. We demonstrate the benefits of the proposed technique using a dataset with 33,126 dermoscopic images from 2056 patients. We evaluate the performance of the proposed technique in terms of the F1-measure, area under the ROC curve (AUC-ROC), and area under the PR-curve (AUC-PR), and compare it with that of seven different benchmark methods, including two recent CNN-based techniques. The proposed technique compares favourably in terms of all the evaluation metrics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Skin cancer is caused by mutations within the DNA of skin cells, which causes their abnormal multiplication [1, 2]. In the early development of skin cancer, lesions appear on the the outer layer of the skin, the epidermis. Not all lesions are caused by malignant tumours, and a diagnosis classifying the lesion as either malignant (cancerous) or benign (non-cancerous) is often reached based on preliminary visual inspection followed by a biopsy. Early detection and classification of lesions is important because early diagnosis of skin cancer significantly improves the prognosis [3].
The visual inspection of potentially malignant lesions carried out using an optical dermatoscope is a challenging task and requires a specialist dermatologist. For instance, according to Morton and Machie [4], in the case of melanoma, a particularly aggressive type of skin cancer, only about 60–90% of malignant tumours are identified based on visual inspection, and accuracy varies markedly depending on the experience of the dermatologist. As skillful dermatologists are not available globally and for all ethnic and socioeconomic groups, the situation causes notable global health inequalities [5].
Due to the aforementioned reasons, machine learning techniques are widely studied in the literature. Machine learning has potential to aid automatic detection of skin cancer from dermoscopic images, thus enabling early diagnosis and treatment. Murugan et al. [6] compared the performance of K-nearest neighbor (KNN), random forest (RF), and support vector machine (SVM) classifiers on data extracted from segmented regions of demoscopic images. Similarly, Ballerini et al. [7] used a KNN-based hierarchical approach for classifying five different types of skin lesions. Thomas et al. [8] used deep learning based methods for classification and segmentation of skin cancer. Lau and Al-Jumaily [9] proposed a technique based on a Multi-Layer Perceptron (MLP) and other neural network models. Chaturvedi et al. [10] used deep CNN for the classification of skin cancer into different classes. Jinnai et al. [11] used region based CNN for efficient detection of skin cancer. Whereas, Nawaz et al. [12] introduced deep learning based skin cancer classification technique. During data pre-processing phase visual appearance of image is improved. After that, output from region based CNN is then provided to fuzzy K mean cluster in order to segment out the cancer affected region. Performance of Nawaz’s technique is checked on different datasets. A recent review by Chan et al. [13] summarizing many of these studies concluded that while many authors reported better sensitivity and specificity than dermatologists, “further validation in prospective clinical trials in more real-world settings is necessary before claiming superiority of algorithm performance over dermatologists.”
What all the aforementioned methods have in common is that they require large amounts of training data in the form of dermoscopic images together with labels indicating the correct diagnosis. Several authors have proposed approaches to reduce the amount of training data required to reach satisfactory classification accuraracy. Hosney et al. [14] describe a method based on transfer learning, which is a way to exploit available training data collected for a different classification task than the one at hand. Hosny’s technique is based on a pre-trained AlexNet network (a specific deep learning architecture proposed by Krizhevsky et al. [15]) that was originally trained to classify images on a commonly used ImageNet dataset, and then adapted to perform skin cancer classification by transfer learning. Similarly, Dorj et al. [16] used a pre-trained AlexNet combined with a SVM classifier. Guo and Yang [17] utilized a ResNet network, another commonly used deep learning architecture by He et al. [18]. Li and Shen [19] use a combination of two deep learning models for the segmentation and classification of skin lesions. Hirano et al. [20] suggested a transfer learning based technique in which hyperspectral data is used for the detection of melanoma using a pre-trained GoogleNet model [21]. The same pre-trained model is used by Kassem et al. [22]. Esteva et al. [23] use a pre-trained Inception V3 model [21].
Another commonly used technique to improve classification accuracy when limited training data is available is ensemble learning [24, 25]. The idea is to combine the output of multiple classifiers, called base-learners, by using their outputs as the input to another model that is called a meta-learner, to obtain a consensus classification. Ensemble learning tends to reduce variability and improve the classification accuracy. Ain et al. [26] proposed Genetic Programming ensemble learning technique for skin cancer classification. In Ain’s approach feature extraction is performed using GP and then extracted feature space is provided to ensemble of classifier for classifying input images into different classes. Mahbod et al. [27] proposed an ensemble based hybrid technique involving pre-trained AlexNet, VGG16 [28], and ResNet models as feature extractors. Latent features extracted from these models are combined using SVM and logistic regression classifiers as meta-learners.
In addition to the shortage of large quantities of labeled training data, many clinical datasets have severe class imbalance: the proportion of positive cases tends to be significantly lower than that of the negative cases, see [29]. This reduces the amount of informative data points and lowers the accuracy of many machine learning techniques, and may create a bias that leads to an unacceptably high number of false negatives when the model is deployed in real-world clinical applications. To deal with the class imbalance issue, most of the previously reported techniques use data augmentation, i.e., oversampling training data from the minority (positive) class and/or undersampling the majority class. This tends to lead to increased computational complexity as the amount of training data is in some cases increased many-fold, and risks losing informative data points due to undersampling. Qin et al. [30] used GAN for generating high quality images, which ultimately improved the performance of deep learning based classifier. Zunair and Hamza [31] proposed technique which comprises of two steps. In the first step, a CycleGAN [32] generative network is trained to do image-to-image translation from negative to positive samples in order to augment the minority (positive) class data. The synthetic data along with the original data samples are then used as the input to a combination of five (out of 16) layers of the VGG16 network, a pooling layer, and a softmax classification layer.
In this paper, we propose a new technique for skin cancer classification from dermoscopic images based on transfer learning and ensembles of deep neural networks. The motivation behind the proposed technique is to maximize the diversity of the base-learners at various levels during the training of the ensemble to improve the overall accuracy. In our proposed ensembled-based technique, a number of CNN base-learners are trained on input images scaled to different sizes between \(32\times 32\) and \(256\times 256\) pixels. During training, two out of six base-learners are pre-trained CNNs trained on another skin cancer dataset that is not part of primary dataset. In the second step, all the predictions from base-learners along with the meta-data, including, e.g., the age and gender of the subject, provided with the input images is provided to an SVM meta-learner to obtain the final classification. By virtue of training the base-classifiers on input images of different sizes, the model is able to focus on features in multiple scales at the same time. The use of meta-data further diversifies the information which improves the classification accuracy.
We evaluate the performance of the proposed technique on data from the International Skin Imaging Collaboration (ISIC) 2020 Challenge, which is highly imbalanced containing less than 2% malignant samples [33]. Our experiments demonstrate that (i) ensemble learning significantly improves the accuracy even though the accuracy of each of the base-learners is relatively low; (ii) transfer learning and the use of meta-data have only a minor effect on the overall accuracy; (iii) overall, the proposed method compares favourably against to all of the other methods in the experiments, even though the differences between the top performing methods fit within a statistical margin of error.
In Sect. 2, we describe the proposed method, including the architecture of the CNN blocks that are used as the base-learners in ensemble, the transfer learning procedure, as well as the SVM meta-classifier. In Sect. 3, we describe the main dataset and its pre-processing and division into training, validation, and test sets; evaluation metrics are also discussed in the same section. Sect. 4 covers the experimental results in terms of accuracy as well as computational complexity, including additional results that characterize the impact of the ensemble and transfer learning on the performance. Limitations of the study and some open research questions suggested by our study are discussed in Sect. 5, followed by conclusions in Sect. 6.
2 The Proposed Method
The proposed technique is an ensemble-based technique in which CNNs are used as base learners. The base learners are either pre-trained on a balanced dataset collected from the ISIC archive or trained directly on the ISIC 2020 dataset, which is our target dataset. Predicted probabilities of the positive class from all the base learners along with the auxiliary data contained in the metadata associated to the images are used as input to an SVM classifier, which is trained to classify each image in the dataset as positive (malignant) or negative (benign). Figure 1 shows the flowchart of the proposed technique.
2.1 Architecture
In the proposed technique, six base-learners are used. Each of the base learners operates on input data of different dimensions. During training four base learners, \(\mathrm {CNN}_{32\times 32}\), \(\mathrm {CNN}_{64\times 64}\), \(\mathrm {CNN}_{128\times 128}\), and \(\mathrm {CNN}_{256\times 256}\), are trained from random initial parameters on \(32\times 32\), \(64\times 64\), \(128\times 128\), and \(256\times 256\) input images respectively. Another two base learners are trained on malignant and benign skin cancer images of sizes \(32\times 32\) and \(64 \times 64\) respectively, which are not part of ISIC 2020 dataset. We used the F1-measure on the validation data to tune the architectures and the other hyperparameters as explained in more detail in Sect. 3 and “Appendix A”.
2.2 Transfer Learning During Training of the Base-Learners
Transfer learning is used to transfer the knowledge extracted from one type of machine learning problem to another [34, 35]. The domain from where information is extracted is known as the source domain, and the domain where extracted information is applied is called the target domain. The benefit of transfer learning is that it not only saves time that is needed to train the network from scratch but also aid in improving the performance in the target domain.
In the proposed technique, the idea of transfer learning is exploited during the training phase of the base-learners. We pre-train some of the CNN base-learners on a balanced dataset collected from the ISIC archive.Footnote 1 This archive dataset was constructed in 2019, so none of the ISIC 2020 data are included in it. The rest of the base-learners are trained on the ISIC 2020 dataset that comprises the target domain. The introduction of the CNNs pre-trained on balanced data provides a diverse set of predictions, complementing the information coming from the base-learners trained on the ISIC 2020 data. Moreover, since the pre-trained base-learners need to be trained only once instead of re-training them every time we repeat the experiment on random subsets of the ISIC 2020 data (see Sect. 3.2 below), the pre-training saves time.
2.3 SVM as a Meta-classifier
In the proposed technique SVM is used as a meta-learner. The predictions from the base learners, in the format of probabilities of the positive class, are fed as input to the SVM along with the metadata. The purpose of using multiple deep learning modules as an ensemble is to ensure that the SVM meta-classifier can benefit from diverse information extracted from the input images by the the different base-learners, along with side information contained in the metadata.
We use a radial basis function (RBF) kernel of degree 2; for further details, see “Appendix A”.
3 Data and Evaluation Metrics
We use the ISIC 2020 Challenge dataset to train and test the proposed method along with a number of benchmark methods and evaluated their performance with commonly used metrics designed for imbalanced data.
3.1 Dataset and Pre-processing
The dataset used in the proposed technique contains 33,126 dermoscopic images collected from 2056 patients [33]. All the images are labelled using histopathology and expert opinion either as benign or malignant skin lesions. The ISIC 2020 dataset also contains another 10982 test data images without the actual labels, but since we are studying the supervised classification task, we use the labeled data only. All the images are in the JPG format of varying dimensions and shape. We use different input dimensions for different base-learners, so we scale the input images to sizes \(32\times 32\), \(64\times 64\), \(128\times 128,\) and \(256\times 256\) pixels.Footnote 2
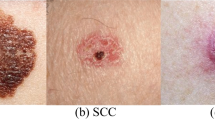
Figure 2 contains example images present in the dataset. Table 1 shows the features in the metadata. Categorical features were encoded as integers in order to reduce the number of parameters in the meta-learner. All the missing values in the metadata are replaced by the average value of the feature in question.
The ISIC 2020 data set is highly imbalanced because out of the total 33,126 images (2056 patients), only 584 images (corresponding to 428 distinct patients) are malignant. The division of the data into training, validation, and test sets is described below in Sect. 3.2.
3.2 Division of the Data and Hyperparameter Tuning
We use the validation set method to divide the data in three parts. As illustrated in Fig. 3, 10% of the total data D is kept as test data, \(D_\mathrm {Test}\), which is not used in the training process. The other 90% is further split by using 90% of it as training data, \(D_\mathrm {T}\), and the final part as validation data, \(D_\mathrm {V}\) which is used to tune the hyperparameters of each of the used methods. Since the ISIC 2020 dataset contains multiple images for the same patient, we require that input images from a given individual appear only in one part of the data (\(D_\mathrm {T}\), \(D_\mathrm {V}\), or \(D_\mathrm {Test}\)).Footnote 3 The validation data is used to adjust the hyperparameters in each of the methods in the experiments by maximizing the F1-score (see Sect. 3.3 below). Hyperparameter tuning was done manually starting from the settings proposed by the original authors (when available) in case of the compared methods, and adjusting them until no further improvement was observed. Likewise, the neural network architectures of the CNN base-learners used in the proposed method were selected based on the same procedure as the other hyperparameters. Tables 5, 6, 7, 8, 9 and 10 in the “Appendix” show the details of the architectures of the CNN base-learners as well as the hyperparameters of the SVM meta-learner.
We used the training and validation data from one random train-validation-test split to tune all hyperparameters. We then used the obtained settings in 10 new random repetitions with independent splits to evaluate the classification performance in order to avoid bias caused by overfitting.
3.3 Evaluation Metrics
As is customary in clinical applications with imbalanced datasets, we use the F1-measure, the area under the ROC curve (AUC-ROC), and the area under the precision–recall curve (AUC-PR) as evaluation metrics; see, e.g., [29]. The F1-measure is the harmonic mean of precision and recall (see definitions below), which is intended to balance the risk of false positives and false negatives. To evaluate the optimal F1-value, we set in each case the classification threshold to the value that maximizes the F1-measure.
AUC-ROC and AUC-PR both characterize the behavior of the classifier over all possible values of the classification threshold. AUC-ROC is the area under the curve between the true positive rate (TPR) and the false positive rate (FPR) at different values of the classification threshold, whereas AUC-PR is the area under the precision–recall curve. While both measures are commonly used in clinical applications, according to Davis and Goadrich [37] and Saito and Rehmsmeier [38], AUC-PR is the preferred metric in cases with imbalanced data where false negatives are of particular concern.
The used metrics are defined as follows:
where \(T_P\) is the number of true positives (positive samples that are correctly classified by the classifier), \(F_P\) is the number of false positives (negative samples incorrectly classified as positive), and P and N are the total number of positive and negative samples, respectively.
4 Experimental Results
For the implementation of the deep learning models we use the Keras version 2.2.4 and TensorFlow version 1.14.0. The other machine learning methods and preprocessing methods were implemented in Python 3.0 and scikit-learn version 0.15.2.
4.1 Computational Cost
We carried out all the experiments on a high-performance computing cluster using maximum four Intel Xeon Gold 5230 CPUs, two Nvidia Volta V100 GPUs, and 300GB of memory for each method. Precise timing comparisons are not straightforward due to variable load on the cluster, but the relative differences are large enough to draw the following qualitative conclusions.
The conventional methods (KNN, RF, MLP, SVM) were the fastest, requiring up to about 1 hour to complete one training and testing cycle. The method proposed by Esteva et al. [23] using an undersampled and augmented version of the ISIC 2020 dataset used 3.1 hours, while our proposed method used 5.2 hours on the full ISIC 2020 dataset. The method by Mahbod et al. [27] was clearly the slowest and took over 72 hours to complete one training and testing cycle even on an undersampled and augmented version of the ISIC 2020 dataset. However, it is worth noting that once the model has been trained, which only needs to be done when the training data is updated, processing new test inputs takes a negligible amount of time compared to the training times (except for the KNN method for which there is no training stage).
4.2 Main Experimental Results
Table 2 and Fig. 4 show a comparison of the proposed technique with four non-deep learning classifiers (KNN, RF, MLP, SVM) and three selected deep learning based techniquesFootnote 4; see “Appendix B” for the most important parameters of the benchmark methods. In each of the benchmark methods except those by Esteva et al. [23] and Mahbod et al. [27], the \(32\times 32\) pixel RGB input images (altogether 3072 input features) along with the auxiliary information in the metadata (additional 3 input features) were used as the input.Footnote 5
In the case of the methods by Esteva et al. [14] and Mahbod et al. [27], we apply downsampling of the majority class to balance the two classes, and use the same data augmentation procedures as described in the original article; for details, see “Appendix B”.
The proposed technique achieves average F1, AUC-PR, and AUC-PR values 0.23, 0.16, and 0.87 respectively, which are highest among all of the compared methods. However, the differences between the top performing methods are within the statistical margin of error.Footnote 6 A more detailed visualization of the ROC and PR-curves is shown in Figs. 5, 6 and 7.
Classification accuracy of the proposed method and seven other methods measured by three evaluation metrics (F1-measure, AUC-PR, AUC-ROC). The scores are averages over \(n=10\) independent repetitions. Error bars are 95% confidence intervals based on the t-distribution with \(n-1=9\) degrees of freedom
4.3 Performance Gain from Ensemble Learning
The proposed technique is comprised of two steps; in the first step base-learners are trained and in the second step meta-learner is trained on the top of base-learners. Table 3 shows the performance of each of the base-learners individually, which can be compared with the performance of the resulting SVM meta-classifier that combines the base-learners outputs as the ensemble classification. Out of six base-learners four are trained from scratch on the ISIC 2020 dataset while the remaining two are pre-trained on skin cancer images that are not part of ISIC 2020 dataset. The performance comparison shows that even though the accuracies of each of the base-learners individually are quite low, the meta-classifier performs markedly better. This suggests that the base-learners succeed in providing a diverse set of inputs to the meta-learner, thus significantly improving the overall performance of the ensemble over any of the base-learners.
4.4 Significance of Transfer Learning and Meta-data
To evaluate the impact of using pre-trained models and that of the metadata on the classification accuracy in the proposed method, we also evaluated the performance with either one of these components disabled. Table 4 shows the performance comparison of the proposed technique to a version where the pre-trained CNNs are disabled, and one where the metadata is not included as auxiliary data for the meta-learner. As seen in the table, excluding the pre-trained CNNs does not significantly affect the performance. The exclusion of the metadata led to somewhat inferior performance, but here too, the differences are relatively minor and is within the statistical margin of the error. Further research with larger datasets and richer metadata is needed to confirm the benefits.
5 Limitations and Open Questions
As with all empirical work, the results from our experiments are subject to various biases, most notably the data set bias, so that the results cannot be directly transferred to other datasets. Further work and experiments on other high-quality, carefully curated data sets is necessary to validate our results. While we have made an effort to avoid overfitting by using a carefully planned training, validation, and testing procedure, the need for architecture optimization and hyperparameter tuning poses challenges to reproducibility. This issue is further amplified by the fluidity of the line between method and preprocessing especially in deep learning, which makes it hard to carry out fair head-to-head comparisons. Further work towards reproducibility standards is acutely needed [40].
Our experimental setup includes only a limited set of baseline and benchmark methods, and it is likely that better performing methods are quite certainly, will be available. The purpose of our work is not only to propose accurate skin cancer detection tools, but more importantly, to study generic techniques that can be used in combination with any existing or future machine learning methods. We hope that they will eventually find their way into clinical work, overcoming some of the current limitations [13] and help in reducing the global health inequalities due to limited availability of qualified dermatologists [5].
Additional open questions and promising research directions include: scaling to massive datasets created by extensive data augmentation; exploring the importance of metadata (on which our results, where the improvement was limited, should not be taken to be conclusive); and the use of enriched imaging data including hyperspectral images [41].
6 Conclusions
We proposed an ensemble-based deep learning approach for skin cancer detection based on dermoscopic images. Our method uses an ensemble of CNNs trained on input images of different sizes along with metadata. We present our results on the ISIC 2020 dataset which contains 33,126 dermoscopic images from 2056 patients. The dataset is highly imbalanced with less than 2% malignant cases. The impact of ensemble learning was found to be significant, while the impact of transfer learning and the use of auxiliary information in the form of metadata associated with the input images appeared to be minor. The proposed method compared favourably against other machine learning based techniques including three deep learning based techniques, making it a promising approach for skin cancer detection especially on imbalanced datasets. Our research expands the evidence suggesting that deep learning techniques offer useful tools in dermatology and other medical applications.
Notes
Some of the raw images are non-square shaped, in which case we reshape them to make the bounding box square and of the desired size.
We use the GroupKFold method in scikit-learn package [36] to do the splitting.
The computational cost of training the SVM classifier prohibited the use of the higher-resolution images.
Following [39], we present comparisons in terms of confidence intervals instead of hypothesis tests (“we argue that the conclusions from comparative classification studies should be based primarily on effect size estimation with confidence intervals, and not on significance tests and p values”). We calculate 95% confidence intervals based on the t-distribution with \(n-1=9\) degrees of freedom by \(\mu \pm 2.26216 \times {\sigma \over \sqrt{n}}\), where \(\mu \) is the average score, \(\sigma \) is the standard deviation of the score, and \(n=10\) is the sample size (number of repetitions with independent random train-validation-test splits).
References
Armstrong BK, Kricker A (1995) Skin cancer. Dermatol Clin 13(3):583–594. https://doi.org/10.1016/S0733-8635(18)30064-0
Simões MF, Sousa JS, Pais AC (2015) Skin cancer and new treatment perspectives: a review. Cancer Lett 357(1):8–42. https://doi.org/10.1016/j.canlet.2014.11.001
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21609
Morton C, Mackie R (1998) Clinical accuracy of the diagnosis of cutaneous malignant melanoma. Br J Dermatol 138(2):283–287. https://doi.org/10.1046/j.1365-2133.1998.02075.x
Buster KJ, Stevens EI, Elmets CA (2012) Dermatologic health disparities. Dermatol Clin 30:53. https://doi.org/10.1016/j.det.2011.08.002
Murugan A, Nair SAH, Kumar KS (2019) Detection of skin cancer using SVM, random forest and kNN classifiers. J Med Syst 43(8):1–9. https://doi.org/10.1007/s10916-019-1400-8
Ballerini L, Fisher RB, Aldridge B, Rees J (2013) A color and texture based hierarchical K-NN approach to the classification of non-melanoma skin lesions. In: Color medical image analysis, pp 63–86. https://doi.org/10.1007/978-94-007-5389-1_4
Thomas SM, Lefevre JG, Baxter G, Hamilton NA (2021) Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer. Med Image Anal 68:101915. https://doi.org/10.1016/j.media.2020.101915
Lau HT, Al-Jumaily A (2009) Automatically early detection of skin cancer: study based on neural network classification. In: 2009 international conference of soft computing and pattern recognition. IEEE, pp 375–380. https://doi.org/10.1109/socpar.2009.80
Chaturvedi SS, Tembhurne JV, Diwan T (2020) A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl 79(39):28477–28498. https://doi.org/10.47611/harp.95
Jinnai S, Yamazaki N, Hirano Y, Sugawara Y, Ohe Y, Hamamoto R (2020) The development of a skin cancer classification system for pigmented skin lesions using deep learning. Biomolecules 10(8):1123. https://doi.org/10.3390/biom10081123
Nawaz M, Mehmood Z, Nazir T, Naqvi RA, Rehman A, Iqbal M, Saba T (2022) Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc Res Tech 85(1):339–351. https://doi.org/10.1002/jemt.23908
Chan S, Reddy V, Myers B, Thibodeaux Q, Brownstone N, Liao W (2020) Machine learning in dermatology: current applications, opportunities, and limitations. Dermatol Ther 10(3):365–386. https://doi.org/10.1007/s13555-020-00372-0
Hosny KM, Kassem MA, Fouad MM (2020) Classification of skin lesions into seven classes using transfer learning with AlexNet. J Digit Imaging 33(5):1325–1334. https://doi.org/10.1007/s10278-020-00371-9
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ (eds) Advances in Neural Information Processing Systems. Morgan Kaufmann Publishers Inc., San Francisco
Dorj U-O, Lee K-K, Choi J-Y, Lee M (2018) The skin cancer classification using deep convolutional neural network. Multimed Tools Appl 77(8):9909–9924. https://doi.org/10.1007/s11042-018-5714-1
Guo S, Yang Z (2018) Multi-Channel-ResNet: an integration framework towards skin lesion analysis. Inform Med Unlocked 12:67–74. https://doi.org/10.1016/j.imu.2018.06.006
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778. https://doi.org/10.1109/cvpr.2016.90
Li Y, Shen L (2018) Skin lesion analysis towards melanoma detection using deep learning network. Sensors 18(2):556. https://doi.org/10.3390/s18020556
Hirano G, Nemoto M, Kimura Y, Kiyohara Y, Koga H, Yamazaki N, Christensen G, Ingvar C, Nielsen K, Nakamura A et al (2020) Automatic diagnosis of melanoma using hyperspectral data and GoogLeNet. Skin Res Technol 26(6):891–897. https://doi.org/10.1111/srt.12891
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: 2016 IEEE conference on computer vision and pattern recognition, CVPR 2016, pp 2818–2826. https://doi.org/10.1109/CVPR.2016.308
Kassem MA, Hosny KM, Fouad MM (2020) Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access 8:114822–114832. https://doi.org/10.1109/access.2020.3003890
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118. https://doi.org/10.1038/nature21056
Dietterich TG et al (2002) Ensemble learning. In: The handbook of brain theory and neural networks, vol 2, pp 110–125. https://doi.org/10.7551/mitpress/3413.003.0009
Polikar R (2012) Ensemble learning. In: Ensemble machine learning, pp 1–34. https://doi.org/10.1007/978-1-4419-9326-7_1
Ain QU, Al-Sahaf H, Xue B, Zhang M (2020) A genetic programming approach to feature construction for ensemble learning in skin cancer detection. In: Proceedings of the 2020 genetic and evolutionary computation conference, pp 1186–1194. https://doi.org/10.1145/3377930.3390228
Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I (2019) Skin lesion classification using hybrid deep neural networks. In: ICASSP 2019–2019 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1229–1233. https://doi.org/10.1109/icassp.2019.8683352
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: Bengio Y, LeCun Y (eds) 3rd international conference on learning representations (ICLR)
He H, Garcia E (2009) Learning from imbalanced data. IEEE Trans Knowl Data Eng 21(9):1263–1284. https://doi.org/10.1109/TKDE.2008.239
Qin Z, Liu Z, Zhu P, Xue Y (2020) A GAN-based image synthesis method for skin lesion classification. Comput Methods Programs Biomed 195:105568. https://doi.org/10.1016/j.cmpb.2020.105568
Zunair H, Hamza AB (2020) Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol 65(13):135005. https://doi.org/10.1088/1361-6560/ab86d3
Zhu J-Y, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision, pp 2223–2232. https://doi.org/10.1109/iccv.2017.244
Rotemberg V, Kurtansky N, Betz-Stablein B, Caffery L, Chousakos E, Codella N, Combalia M, Dusza S, Guitera P, Gutman D et al (2021) A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data 8(1):1–8. https://doi.org/10.1038/s41597-021-00815-z
Torrey L, Shavlik J (2010) Transfer learning. In: Handbook of research on machine learning applications and trends algorithms, methods, and techniques, pp 242–264. https://doi.org/10.4018/978-1-60566-766-9.ch011
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/tkde.2009.191
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine Learning in Python. J Mach Learn Res 12:2825–2830
Davis J, Goadrich M (2006) The relationship between precision–recall and ROC curves. In: Proceedings of the 23rd international conference on machine learning, pp 233–240. https://doi.org/10.1145/1143844.1143874
Saito T, Rehmsmeier M (2015) The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 10:0118432. https://doi.org/10.1371/journal.pone.0118432
Berrar D, Lozano JA (2013) Significance tests or confidence intervals: which are preferable for the comparison of classifiers? J Exp Theor Artif Intell 25:189–206. https://doi.org/10.1080/0952813X.2012.680252
Heil BJ, Hoffman MM, Markowetz F, Lee S-I, Greene CS, Hicks SC (2021) Reproducibility standards for machine learning in the life sciences. Nat Methods 18:1132–1135. https://doi.org/10.1038/s41592-021-01256-7
Johansen TH, Møllersen K, Ortega S, Fabelo H, Garcia A, Callico GM, Godtliebsen F (2020) Recent advances in hyperspectral imaging for melanoma detection. WIREs Comput Stat 12(1):1465. https://doi.org/10.1002/wics.1465
Acknowledgements
This work was funded in part by the Academy of Finland (Project TensorML #311277).
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Detailed Architecture and Hyperparameters of the Proposed Method
See Tables 5, 6, 7, 8, 9, 10 and 11.
Appendix B: Data Preprossing and Hyperparameters of the Compared Methods
We preprocessed the data to optimize the performance under reasonable computational limits of each of the used methods separately, following the original preprocessing and data augmentation protocols proposed in the original articles where applicable. In case of all the baseline classifiers (KNN, RF, MLP, SVM) all the input images are resized to \(32\times 32\) size and normalized. For the Mahbod et al. [27] technique, we undersampled the majority class to achieve better balance between the positive and negative classes, and performed data augmentation as described in the original article. The input images were rescaled to match the input dimension of the three base-learners (AlexNet, Resnet and VGG16). For the Esteva et al. [23] technique, the input images were resized to match the input dimension of VGG16, the training data was undersampled to balance the classes, and 4x data augmentation by rotation and mirroring was performed in the way described in the original article. In all cases, all the positive samples were retained.
We used the following hyperparameters for the compared methods, which were selected manually by adjusting them until no further improvement on the validation data performance (F1-measure) was observed. For the KNN classifier, we use \(k=4\). For the random forest (RF), maximum depth 30 and \(n=100\) trees are used. For the multilayer perceptron (MLP), we use one hidden layer with 120 neurons, and minibatch size 250. For the support vector machine (SVM), a polynomial kernel of degree 3, and constants \(C=0.07\) and \(\gamma =0.0009\) are used. In the deep autoencoder (deep-AE), we use an encoder with two layers having 2000 and 1000 neurons, respectively, and a symmetric decoder, and train the model for 100 epochs with minibatch size 15.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qureshi, A.S., Roos, T. Transfer Learning with Ensembles of Deep Neural Networks for Skin Cancer Detection in Imbalanced Data Sets. Neural Process Lett 55, 4461–4479 (2023). https://doi.org/10.1007/s11063-022-11049-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-022-11049-4