Abstract
Purpose
Ependymoma (EPN) accounts for approximately 10% of all primary central nervous system (CNS) tumors in children and in most cases, chemotherapy is ineffective and treatment remains challenging. We investigated molecular alterations, with a potential prognostic marker and therapeutic target in EPNs of childhood and adolescence, using a next-generation sequencing (NGS) panel specific for pediatric neoplasms.
Methods
We selected 61 samples with initial diagnosis of EPN from patients treated at Pediatric Oncology Institute-GRAACC/UNIFESP. All samples were divided according to the anatomical compartment of the CNS - 42 posterior fossa (PF), 14 supratentorial (ST), and five spinal (SP). NGS was performed to identify somatic genetic variants in tumor samples using the Oncomine Childhood Cancer Research Assay® (OCCRA®) panel, from Thermo Fisher Scientific®.
Results
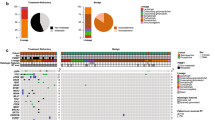
Genetic variants were identified in 24 of 61 (39.3%) tumors and over 90% of all variants were pathogenic or likely pathogenic. The most commonly variants detected were in CIC, ASXL1, and JAK2 genes and have not been reported in EPN yet. MN1-BEND2 fusion, alteration recently described in a new CNS tumor type, was identified in one ST sample that was reclassified as astroblastoma. Additionally, YAP1‐MAMLD1 fusion, a rare event associated with good outcome in ST-EPN, was observed in two patients diagnosed under 2 years old.
Conclusions
Molecular profiling by the OCCRA® panel showed novel alterations in pediatric and adolescent EPNs, which highlights the clinical importance in identifying genetic variants for patients’ prognosis and therapeutic orientation.



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C (2019) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol 21:v1–v100. https://doi.org/10.1093/neuonc/noz150
Jünger ST, Timmermann B, PietschT, (2021) Pediatric ependymoma: an overview of a complex disease. Childs Nerv Syst 37:2451–2463. https://doi.org/10.1007/s00381-021-05207-7
Khatua S, Mangum R, Bertrand KC, Zaky W, McCall D, Mack SC (2018) Pediatric ependymoma: current treatment and newer therapeutic insights. Future Oncol 14(30):3175–3186. https://doi.org/10.2217/fon-2018-0502
Pajtler KW, Witt H, Sill M et al (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27(5):728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Pajtler KW, Mack SC, Ramaswamy V et al (2017) The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol 133(1):5–12. https://doi.org/10.1007/s00401-016-1643-0
Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251. https://doi.org/10.1093/neuonc/noab106
Arabzade A, Zhao Y, Varadharajan S et al (2021) ZFTA-RELA dictates oncogenic transcriptional programs to drive aggressive supratentorial ependymoma. Cancer Discov 1066:2020. https://doi.org/10.1158/2159-8290.CD-20-1066
Zhu JJ, Jillette N, Li XN, Cheng AW, Lau CC (2020) C11orf95-RELA reprograms 3D epigenome in supratentorial ependymoma. Acta Neuropathol 140(6):951–960. https://doi.org/10.1007/s00401-020-02225-8
Jünger ST, Timmermann B, Pietsch T (2021) Pediatric ependymoma: an overview of a complex disease. Childs Nerv Syst. https://doi.org/10.1007/s00381-021-05207-7
Witt H et al (2011) Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20(2):143–157. https://doi.org/10.1016/j.ccr.2011.07.007
Gajjar A, Pfister SM, Taylor MD, Gilbertson RJ (2014) Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin Cancer Res 20(22):5630–5640. https://doi.org/10.1158/1078-0432.CCR-14-0833
Kline CN et al (2017) Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol 19(5):699–709. https://doi.org/10.1093/neuonc/now254
Ahmed AA, Vundamati DS, Farooqi MS, Guest E (2018) Precision medicine in pediatric cancer: current applications and future prospects. High Throughput 7(4):39. https://doi.org/10.3390/ht7040039
Hiemenz MC, Ostrow DG, Busse TM et al (2018) OncoKids: a comprehensive next-generation sequencing panel for pediatric malignancies. J Mol Diagn 20(6):765–776. https://doi.org/10.1016/j.jmoldx.2018.06.009
Wang L, Liu L, Li H et al (2019) RELA fusion in supratentorial extraventricular ependymomas: a morphologic, immunohistochemical, and molecular study of 43 cases. Am J Surg Pathol 43(12):1674–1681. https://doi.org/10.1097/PAS.0000000000001342
Mack SC, Witt H, Piro RM et al (2014) Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506(7489):445–450. https://doi.org/10.1038/nature13108
van Veelen-Vincent ML, Pierre-Kahn A, Kalifa C et al (2002) Ependymoma in childhood: prognostic factors, extent of surgery, and adjuvant therapy. J Neurosurg 97(4):827–835. https://doi.org/10.3171/jns.2002.97.4.0827
Cho HJ, Park HY, Kim K et al (2021) Methylation and molecular profiles of ependymoma: influence of patient age and tumor anatomic location. Mol Clin Oncol 14(5):88. https://doi.org/10.3892/mco.2021.2250
Pagès M, Pajtler KW, Puget S et al (2019) Diagnostics of pediatric supratentorial RELA ependymomas: integration of information from histopathology, genetics, DNA methylation and imaging. Brain Pathol 29(3):325–335. https://doi.org/10.1111/bpa.12664
Jungwirth G et al (2019) Intraventricular meningiomas frequently harbor NF2 mutations but lack common genetic alterations in TRAF7, AKT1, SMO, KLF4, PIK3CA, and TERT. Acta Neuropathol Commun 7(1):140. https://doi.org/10.1186/s40478-019-0793-4
Shankar GM et al (2014) Sporadic hemangioblastomas are characterized by cryptic VHL inactivation. Acta Neuropathol Commun 2:167. https://doi.org/10.1186/s40478-014-0167-x
Bunda S, Heir P, Metcalf J et al (2019) CIC protein instability contributes to tumorigenesis in glioblastoma. Nat Commun 10:661. https://doi.org/10.1038/s41467-018-08087-9
Jiménez G, Shvartsman SY, Paroush Z (2012) The Capicua repressor–a general sensor of RTK signaling in development and disease. J Cell Sci 125(Pt6):1383–1391. https://doi.org/10.1242/jcs.092965
Liu Z, Liu H, Liu Z, Zhang J (2015) Oligodendroglial tumours: subventricular zone involvement and seizure history are associated with CIC mutation status. BMC Neurol 19(1):134. https://doi.org/10.1186/s12883-019-1362-y
Cahill DP, Louis DN, Cairncross JG (2015) Molecular background of oligodendroglioma: 1p/19q, IDH, TERT, CIC and FUBP1. CNS Oncol 4(5):287–294. https://doi.org/10.2217/cns.15.32
Yip S et al (2012) Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 226(1):7–16. https://doi.org/10.1002/path.2995
Yang R et al (2017) CIC loss promotes gliomagenesis via aberrant neural stem cell proliferation and differentiation. Can Res 77(22):6097–6108. https://doi.org/10.1158/0008-5472.CAN-17-1018
Weissmann S et al (2018) The tumor suppressor CIC directly regulates MAPK pathway genes via histone deacetylation. Can Res 78(15):4114–4125. https://doi.org/10.1158/0008-5472.CAN-18-0342
Simón-Carrasco L, Jiménez G, Barbacid M, Drosten M (2018) The Capicua tumor suppressor: a gatekeeper of Ras signaling in development and cancer. Cell Cycle 17(6):702–711. https://doi.org/10.1080/15384101.2018.1450029
Inoue D, Fujino T, Kitamura T (2018) ASXL1 as a critical regulator of epigenetic marks and therapeutic potential of mutated cells. Oncotarget 9(81):35203–35204. https://doi.org/10.18632/oncotarget.26230
Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D (2012) Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol 5:12. https://doi.org/10.1186/1756-8722-5-12
Lulla RR, Saratsis AM, Hashizume R (2016) Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv 2(3):e1501354–e1501354. https://doi.org/10.1126/sciadv.1501354
Szathmari A, Zerah M, Vinchon M, Dufour C, Gimbert E, Di Rocco F, Chabaud S, Conter C, Mottolese C, Frappaz D (2019) Ependymoma of the Spinal Cord in children: a retrospective French study. World Neurosurg 126:e1035–e1041. https://doi.org/10.1016/j.wneu.2019.03.033
Lin Y, Jea A, Melkonian SC, Lam S (2015) Treatment of pediatric grade II spinal ependymomas: a population-based study. J Neurosurg Pediatr 15(3):243–249. https://doi.org/10.3171/2014.9.PEDS1473
Phi JH et al (2015) Overcoming chemoresistance of pediatric ependymoma by inhibition of STAT3 signaling. Transl Oncol 8(5):376–386. https://doi.org/10.1016/j.tranon.2015.08.001
Mukthavaram R et al (2015) Effect of the JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres. J Transl Med 13:269. https://doi.org/10.1186/s12967-015-0627-5
Lin M et al (2019) JAK2 p.G571S in B-cell precursor acute lymphoblastic leukemia: a synergizing germline susceptibility. Leukemia 33(9):2331–2335. https://doi.org/10.1038/s41375-019-0459-z
Sturm D, Orr BA, Toprak UH et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164(5):1060–1072. https://doi.org/10.1016/j.cell.2016.01.015
Burford A, Mackay A, Popov S et al (2018) The ten-year evolutionary trajectory of a highly recurrent paediatric high grade neuroepithelial tumour with MN1:BEND2 fusion. Sci Rep 8(1):1032. https://doi.org/10.1038/s41598-018-19389-9
Yamasaki K, Nakano Y, Nobusawa S, Okuhiro Y, Fukushima H, Inoue T, Murakami C, Hirato J, Kunihiro N, Matsusaka Y, Honda-Kitahara M, Ozawa T, Shiraishi K, Kohno T, Ichimura K, Hara J (2020) Spinal cord astroblastoma with an EWSR1-BEND2 fusion classified as a high-grade neuroepithelial tumour with MN1 alteration. Neuropathol Appl Neurobiol 46(2):190–193. https://doi.org/10.1111/nan.12593
Pajtler KW et al (2019) YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat Commun 10(1):3914. https://doi.org/10.1038/s41467-019-11884-5
Andreiuolo F, Varlet P, Tauziède-Espariat A et al (2019) Childhood supratentorial ependymomas with YAP1-MAMLD1 fusion: an entity with characteristic clinical, radiological, cytogenetic and histopathological features. Brain Pathol 29(2):205–216. https://doi.org/10.1111/bpa.12659
de Sousa GR, Lira RCP, de Almeida MT et al (2021) A coordinated approach for the assessment of molecular subgroups in pediatric ependymomas using low-cost methods. J Mol Med. https://doi.org/10.1007/s00109-021-02074-2.10.1007/s00109-021-02074-2
Acknowledgements
This study was conducted as a part of the “Investigation of genetic alterations of childhood and adolescence ependymomas and gliomas using the next-generation sequencing strategy” research project, supported by Fundação de Amparo à Pesquisa do Estado de Sao Paulo (The Sao Paulo Research Foundation - FAPESP), and Pediatric Oncology Institute-Grupo de Apoio ao Adolescente e à Criança com Câncer/Federal University of Sao Paulo (IOP-GRAACC/UNIFESP). The authors thank to all the patients and families who contributed to this study.
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de Sao Paulo (FAPESP No. 2019/12074-5), Pediatric Oncology Institute-Grupo de Apoio ao Adolescente e à Criança com Câncer/Federal University of Sao Paulo (IOP-GRAACC/UNIFESP), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Contributions
Conception/design and development of methodology: SRCT, FT, IDO; acquisition of data: DCCC, FT, IDO, SRCT; analysis and interpretation of data: DCCC, FT, IDO, SRCT, NSS, AMC, FABS; writing of manuscript: DCCC; review and/or revision of manuscript: SRCT, FT, NSS, AMC; medical support: NSS, AMC, MTSA, FABS, PAD, SC.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Research Committee (Committee for Ethics in Research – Federal University of Sao Paulo No. 0915/2019). This article does not contain any studies with animals performed by any of the authors.
Informed consent
Samples from each primary tumor were collected after informed consent was signed by patients/guardians. The biological material is acquired via a Biobank of the Pediatric Oncology Institute-GRAACC/UNIFESP (National Commission of Ethics in Research - CONEP B-053).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cabral de Carvalho Corrêa, D., Tesser-Gamba, F., Dias Oliveira, I. et al. Molecular profiling of pediatric and adolescent ependymomas: identification of genetic variants using a next-generation sequencing panel. J Neurooncol 155, 13–23 (2021). https://doi.org/10.1007/s11060-021-03848-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-021-03848-x