Abstract
Background
Peucedani Radix, also known as “Qian-hu” is a traditional Chinese medicine derived from Peucedanum praeruptorum Dunn. It is widely utilized for treating wind-heat colds and coughs accompanied by excessive phlegm. However, due to morphological similarities, limited resources, and heightened market demand, numerous substitutes and adulterants of Peucedani Radix have emerged within the herbal medicine market. Moreover, Peucedani Radix is typically dried and sliced for sale, rendering traditional identification methods challenging.
Materials and methods
We initially examined and compared 104 commercial “Qian-hu” samples from various Chinese medicinal markets and 44 species representing genuine, adulterants or substitutes, utilizing the mini barcode ITS2 region to elucidate the botanical origins of the commercial “Qian-hu”. The nucleotide signature specific to Peucedani Radix was subsequently developed by analyzing the polymorphic sites within the aligned ITS2 sequences.
Results
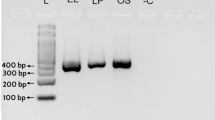
The results demonstrated a success rate of 100% and 93.3% for DNA extraction and PCR amplification, respectively. Forty-five samples were authentic “Qian-hu”, while the remaining samples were all adulterants, originating from nine distinct species. Peucedani Radix, its substitutes, and adulterants were successfully identified based on the neighbor-joining tree. The 24-bp nucleotide signature (5′-ATTGTCGTACGAATCCTCGTCGTC-3′) revealed distinct differences between Peucedani Radix and its common substitutes and adulterants. The newly designed specific primers (PR-F/PR-R) can amplify the nucleotide signature region from commercial samples and processed materials with severe DNA degradation.
Conclusions
We advocate for the utilization of ITS2 and nucleotide signature for the rapid and precise identification of herbal medicines and their adulterants to regulate the Chinese herbal medicine industry.




Similar content being viewed by others
Data availability
The sequences of each haplotype in this study have been submitted to GenBank of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov; see Table S1). Voucher samples were deposited in the School of Pharmaceutical Sciences and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University (Yunnan, China), and detailed information was listed in Table S1.
Abbreviations
- ITS2:
-
Internal transcribed spacer 2
- NJ:
-
Neighbor-joining
- CTAB:
-
Hexadecyltrimethylammonium bromide
- HMM:
-
Hidden Markov model
- MEGA:
-
Molecular evolutionary genetics analysis
- NCBI:
-
National Centre for Biotechnology Information
- K2P:
-
Kimura-2-parameter
References
Shan RH, Sheh ML (1979) Apiaceae. In: Shan RH, Sheh ML (eds) Flora reipublicae popularis sinicae. Science Press, Beijing, pp 13–62
Pimenov MG (2017) Updated checklist of Chinese Umbelliferae: nomenclature, synonymy, typification, distribution. Turczaninowia 20:106–239. https://doi.org/10.14258/turczaninowia.20.2.9
Chinese Pharmacopoeia Commission (2020) Pharmacopoeia of the People’s Republic of China. China Medical Science and Technology Press, Beijing
Wu YS, Wu YY, Liu YJ et al (2017) Advances in Peucedani Radix and its adulterants and substitutes in Chinese materia medica identification. Chin Tradit Herbal Drugs 48:2335–2339
Xu X, Li HF (2016) An analysis of the medicinal value of Peucedanum japonicum Thunb. Asia-Pac Tradit Med 12:45–47
Rao GX, Liu QX, Dai ZJ et al (1995) Textual research for the traditional Chinese medicine radix peucedani and discussion of its modern varieties. J Yunnan Coll Tradit Chin Med 18:1–6
Mishra P, Kumar A, Nagireddy A et al (2016) DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol J 14:8–21. https://doi.org/10.1111/pbi.12419
Hebert PD, Gregory TR (2005) The promise of DNA barcoding for taxonomy. Syst Biol 54:852–859. https://doi.org/10.1080/10635150500354886
Hebert PD, Cywinska A, Ball SL et al (2003) Biological identifications through DNA barcodes. Proc Biol Sci 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Wang CY, Guo BL, Xiao PG (2011) Review on molecular identification methods of traditional Chinese medicine. Chin J Chin Mater Med 36:237–242
Chase MW, Cowan RS, Hollingsworth PM et al (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56:295–299. https://doi.org/10.1002/tax.562004
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508. https://doi.org/10.1371/journal.pone.0000508
Liu ZW, Gao YZ, Zhou J (2019) Molecular authentication of the medicinal species of Ligusticum (Ligustici Rhizoma et Radix, “Gao-ben”) by integrating non-coding internal transcribed spacer 2 (ITS2) and its secondary structure. Front Plant Sci 10:429. https://doi.org/10.3389/fpls.2019.00429
Chen ZY, Lai CJS, Wei XY et al (2020) Research progress on identification methods of growth years of traditional Chinese medicinal materials. Chin J Chin Mater Med 46:1357–1367
Zhao XY, Liu R, Feng H et al (2021) DNA barcoding molecular identification of Platycodon grandiflorum and its confusability. J Hebei Univ 41:60–67
Chen SL, Yao H, Han JP et al (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613. https://doi.org/10.1371/journal.pone.0008613
China Plant BOL Group, Li DZ, Gao LM et al (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108:19641–19646. https://doi.org/10.1073/pnas.1104551108
Zhou J, Wang WC, Liu MQ et al (2014) Molecular authentication of the traditional medicinal plant Peucedanum praeruptorum and its substitutes and adulterants by DNA-barcoding technique. Pharmacogn Mag 10:385–390. https://doi.org/10.4103/0973-1296.141754
Hajibabaei M, Smith MA, Janzen DH et al (2006) A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes 6:959–964. https://doi.org/10.1111/j.1471-8286.2006.01470.x
Wandeler P, Hoeck PE, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642. https://doi.org/10.1016/j.tree.2007.08.017
Lo YT, Li M, Shaw PC (2015) Identification of constituent herbs in ginseng decoctions by DNA markers. Chin Med 10:1–8. https://doi.org/10.1186/s13020-015-0029-x
Liu Y, Wang XY, Wang LL et al (2016) A nucleotide signature for the identification of American ginseng and its products. Front Plant Sci 7:319. https://doi.org/10.3389/fpls.2016.00319
Wang XY, Liu Y, Wang LL et al (2016) A nucleotide signature for the identification of Angelicae Sinensis Radix (Danggui) and its products. Sci Rep 6:1–8. https://doi.org/10.1038/srep34940
Zhang TY, Xu FS, Ruhsam MK et al (2022) A nucleotide signature for the identification of Pinelliae Rhizoma (Banxia) and its products. Mol Biol Rep 49:7753–7763. https://doi.org/10.1007/s11033-022-07600-0
Liu Y (2018) Study of TCM nucleotide signature based on mini-barcoding-Panax quinquefolius, Ophiocordyceps sinensis and Ginkgo biloba for example. Dissertation, Peking Union Medical College
Chen SL, Yao H, Han JP et al (2013) Guidelines for molecular identification of DNA barcoding in Chinese medicinal materials. Chin J Chin Mater Med 38:141–148
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf issue. Phytochem Bull 19:11–15
White TJ, Bruns T, Lee S et al (1999) Amplification and direct sequencing of fungal ribosomal RNA genes for polygenetics. PCR Protocols 18:315–322
Keller A, Schleicher T, Schultz J et al (2009) 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene 430:50–57. https://doi.org/10.1016/j.gene.2008.10.012
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Koetschan C, Hackl T, Müller T et al (2012) ITS2 database IV: interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol Phylogenet Evol 63:585–588. https://doi.org/10.1016/j.ympev.2012.01.026
Zhou J, Gao YZ, Wei J et al (2020) Molecular phylogenetics of Ligusticum (Apiaceae) based on nrDNA ITS sequences: rampant polyphyly, placement of the Chinese endemic species, and a much-reduced circumscription of the genus. Int J Plant Sci 181:306–323. https://doi.org/10.1086/706851
Chinese Pharmacopoeia Commission (1995) Pharmacopoeia of the People’s Republic of China. Guangdong Science and Technology Press, Guangzhou
Liu YL, Fu S, Fan LJ et al (2017) Review on differences in pharmacological, constituents and other aspects between Schisandrae Chinensis Fructus and Schisandrae Sphenantherae Fructus. Chin J Exp Tradit Med Formul 23:228–234
Chinese Pharmacopoeia Commission (2000) Pharmacopoeia of the People’s Republic of China. Chemical Industry Publishing House, Beijing
Zhang C, Xiao YQ, Taniguchi M (2005) Studies on chemical constituents in roots of Peucedanum praeruptorum (I). Chin J Chin Mater Med 30:675–677
Zhang C, Xiao YQ, Taniguchi M (2006) Studies on chemical constituents in roots of Peucedanum praeruptorum (II). Chin J Chin Mater Med 31:1333–1335
Chinese Pharmacopoeia Commission (2015) Pharmacopoeia of the People’s Republic of China. China Medical Science and Technology Press, Beijing
Gansu Food and Drug Administration (2008) Gansu Province Chinese herbal medicine standard. Gansu Culture Publishing House, Lanzhou
Sichuan Food and Drug Administration (2010) Sichuan Province Chinese herbal medicine standard. Sichuan Science and Technology Press, Chengdu
He Y (2021) Identification method and quality analysis of commercial “Qian-hu”. Dissertation, University of Chinese Medicine
Pu FD, Watson MF (2005) Ligusticum L. In: Committee E (ed) Flora of China. Science Press, Beijing, pp 140–150
Su J (1981) Tang materia medica. Anhui Science and Technology Publishing House, Hefei
Zhang HY (1994) Chinese traditional medicine resources. Science Press, Beijing
Chen CB, Wang DR (2006) Commonly used botanical medicines of the Tujia People in Hubei Province (Umbelliferae). Chin J Folk Med 6:354–356
Xue CY, Li DZ (2011) Use of DNA barcode to identify traditional Tibetan medicinal plant Gentianopsis paludosa (Gentianaceae). J Syst Evol 49:267–270. https://doi.org/10.1111/j.1759-6831.2011.00127.x
Gao ZT, Liu Y, Wang XY et al (2019) DNA mini-barcoding: a derived barcoding method for herbal molecular identification. Front Plant Sci 10:987. https://doi.org/10.3389/fpls.2019.00987
Song M, Dong GQ, Zhang YQ et al (2017) Identification of processed Chinese medicinal materials using DNA mini-barcoding. Chin J Nat Med 15:481–486. https://doi.org/10.1016/S1875-5364(17)30073-0
Särkinen T, Staats M, Richardson JE et al (2012) How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS ONE 7:e43808. https://doi.org/10.1371/journal.pone.0043808
Sheh ML, Watson MF (2005) Peucedanum L. In: Committee E (ed) Flora of China. Science Press, Beijing, pp 182–192
Acknowledgements
We thank two anonymous reviewers for valuable, constructive comments that helped improve the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31960048), the Ten Thousand Talents Program of Yunnan (YNWRQNBJ-2019-208), the Department of Science and Technology of Yunnan Province (Grant No. 202201AT070118) and Gaoligong Mountain, Forest Ecosystem, Observation and Research Station of Yunnan Province (Grant No. 202205AM070006).
Author information
Authors and Affiliations
Contributions
ZL and JZ conceived and designed the study, as well as revised the manuscript. JN, XW, JY, and SZ collected samples and conducted the experiments. JN analyzed the data and authored the manuscript. All authors have reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The collection of samples and use comply with relevant institutional, national, and international guidelines and legislation.
Consent to participate
This article does not contain any studies with human participants or animals and did not involve any endangered or protected species.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Niu, J., Wang, X., Zhou, S. et al. Molecular authentication of commercial “Qian-hu” through the integration of nrDNA internal transcribed spacer 2 and nucleotide signature. Mol Biol Rep 51, 639 (2024). https://doi.org/10.1007/s11033-024-09557-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09557-8