Abstract
Background
The radix of Glehnia littoralis Fr. Schmidt ex Miq. (Beishashen), is often misidentified and adultered in Chinese medicine. Its seven common adulterants include Chuanminshen violaceum Sheh et Shan (Chuanmingshen), Changium smyrnioides Wolff (Mingdangshen), Sphallerocarpus gracilis (Bess.) K.-Pol. (Miguoqin), Adenophora polyantha Nakai (Shishashen), Silene tatarinowii Regel (Shishengyingzicao), Adenophora tetraphylla (Thunb.) Fisch (Lunyeshashen) and Adenophora stricta Miq. (Shashen). This study aims to evaluate the feasibility of the second internal transcribed spacer (ITS2) DNA barcoding to discriminate between Glehniae Radix and its common adulterants.
Methods
In this study, we collected 46 samples of G. littoralis and 59 samples of its seven common adulterants. Genomic DNA sequences were extracted from samples, including original plants and commercially processed crude drugs. The ITS2 of the ribosomal DNA sequences were amplified and sequenced bi-directionally. The sequences were assembled by CodonCode Aligner 3.5.7. The descriptive data analysis was conducted and neighbor-joining (NJ) phylogenetic tree was constructed by MEGA 5.0 in accordance with the kimura 2 -parameter (K2P) model. The identification efficiency was evaluated based on the BLAST1 methods. The ITS2 secondary structures were predicted and compared between Glehniae Radix and its adulterants by the ITS2 database.
Results
As the 46 ITS2 sequences of G. littoralis were identical to each other, the identification efficiency of the ITS2 region was 100 %. A NJ tree based on the ITS2 sequences, and the predicted secondary structures of ITS2, distinguished Glehniae Radix from its adulterants.
Conclusion
DNA barcoding based on ITS2 distinguished commercial processed Glehniae Radix from common herbal adulterants.
Similar content being viewed by others
Background
The radix of Glehnia littoralis (beishashen) is used as an antitussive, mucolytic, antibacterial, antiphlogistic and immune response enhancer in Chinese medicine (CM) [1]. It is also used as a diaphoretic, antipyretic and analgesic in Japan [2]. Glehniae Radix is listed in the Japanese and Chinese Pharmacopoeia and is widely recognized as a nutritional and healthy food [3–6].
Due to the high market demand, overexploitation of Glehniae Radix already threatened the existence of this wild species. And it was protected now by China Plants Red Data Book [7]. However, the scarcity of this wild species has resulted in frequent fraudulent adulteration and substitution of G. littoralis with the species Chuanminshen violaceum Sheh et Shan (Chuanmingshen), Changium smyrnioides Wolff (Mingdangshen), Sphallerocarpus gracilis (Bess.) K.-Pol. (Miguoqin), Adenophora polyantha Nakai (Shishashen) and Silene tatarinowii Regel (Shishengyingzicao), because of their similar appearances [8, 9]. Due to their similar Chinese names, Adenophorae Radix is also easily confused with Glehniae Radix in clinical use [10]. The botanical origins of Adenophorae Radix are two species in the family Campanulaceae, Adenophora tetraphylla (Thunb.) Fisch (Lunyeshashen) and Adenophora stricta Miq. (Shashen).
The biologically active compounds of these adulterants are significantly distinct from those of Glehniae Radix. However these adulterants do not contain these biologically active compounds coumarins, coumarin glyeosides and polyacetylenes as Glehniae Radix [11]. The identification of Glehniae Radix and its adulterants has been based on morphological and microscopic observation [8, 9], while molecular identification has been rarely used [10, 12]. However, the assessment procedures are affected by environmental factors and often produce ambiguous results [13].
The DNA barcoding technique uses standard genomic regions to discriminate species [14–16], and this method provides consistent and reliable results regardless of the age, plant part or environmental factors of the sample [17]. Because of its speed and accuracy, DNA barcoding has gained attention in CM identification [12]. Although the Barcode of Life Plant Working Group (BOL) recommends the sequence combination rbcL + matK for barcoding [16], other genomic regions such as nuclear ITS (Internal Transcibed Spacer) may also be useful for medicinal material identification [17]. The ITS2 DNA sequence was suggested as a universal (medicinal) plant barcode [18–20]. The China Plant BOL Group (CBOL) has also suggested that ITS/ITS2 should be incorporated into the core barcode for seed plants [21]. The ITS2 region has been successfully applied to identify diverse medicinal plants and herbal materials [18, 22–26]. However, the ITS2 barcode was more likely to be affected by genetic anomalies, such as gene multiplication, pseudogenes and introgression [20].
For our study, we would like to identify commercial processed Glehniae Radix and its adulterants from a large pool of samples by the ITS2 sequences. This study aims to evaluate the suitability and feasibility of ITS2 barcode to accurately discriminate between Glehniae Radix and its adulterants, particularly the sequence divergences and differentiation powers of the ITS2 region.
Methods
Sample collection
Seven original plant samples (dried leaves or dried roots prepared from plants) and 36 commercially processed crude drug samples belonging to G. littoralis were collected from a large geographical area in China, including Hebei, Shandong and Inner Mongolia (Table 1). We also gathered 16 samples belonging to seven common adulterant species of Glehniae Radix: C. smyrnioides, C. violaceum, A. polyantha, A. tetraphylla, A. stricta, S. gracilis and S. tatarinowii. All samples were identified by Associate Professor Hongxiao Yang (College of Resources and Environment, Qingdao Agricultural University, Qingdao, China) by microscopic and morphological identification [1, 27]. Voucher specimens (Table 1) were deposited in the Herbarium of Qingdao Agricultural University. Silica gel-dried leaves or roots from individual plants were collected and commercially available crude preparations of Glehniae Radix and its adulterants were purchased from pharmacies. Subsequently, three ITS2 sequences of G. littoralis and 43 ITS2 sequences of its seven adulterants were all downloaded from GenBank for further analysis (Table 1).
DNA extraction, amplification and sequencing
Genomic DNA was extracted from the silica gel-dried leaves, dried roots and crude drugs by the plant Genomic DNA Kit (Tiangen BioTech, Beijing, China). DNA extraction from crude drugs required the following improvements. After wiping the treated surface of the samples with 75 % ethanol (Sigma-Aldrich, USA) approximately 150 mg of interior material was obtained. Polyvinyl pyrrolidone (PVP)-30 powder (10 %, Sigma-Aldrich, USA) was added and the material was quickly ground into powder in liquid nitrogen. The process step below was repeated three times. Cold nuclear separation liquid (1 mL; 10 mmol/L Tris–HCl; pH 8.0) (Sigma-Aldrich, USA), 0.3 mol/L saccharose liquid (Sigma-Aldrich, USA), 0.4 % β-mercaptoethanol (Sigma-Aldrich, USA) was added to the powder. The mixture was allowed to stand for 2 min and then centrifuged (Eppendorf, Hamburg, Germany) at 13,500×g for 4 min at 4 °C. The supernatant was then discarded. Next, the precipitate was supplemented with GP1 and incubated overnight in a 65 °C water bath. GP2 was replaced with isopropyl alcohol (Sigma-Aldrich, USA). Further procedures were conducted according to the manufacturer’s instructions. We then performed a PCR amplification of ITS2 by the same primers and PCR conditions as used in previous studies [18]. The extracted DNA and PCR products were examined by 1.0 % agarose gel electrophoresis and scanned by spectrophotometer measurement (Bio-Rad, CA, USA). The purified PCR products were sequenced in both directions on a 3730XL sequencer (Invitrogen BioTech Co. Ltd., Beijing, China).
Data analysis
Consensus and contiguous sequences were generated by a CodonCode Aligner V3.5.7 (CodonCode Co., MA, USA). Then, the ITS2 spacer sequences were obtained after removal of both the 5.8S and 28S sections of the sequences based on Hidden Markov models [28]. The obtained ITS2 sequences were shown to be reliable by the BLAST1 method. Subsequently, K2P genetic distances were calculated by MEGA 5.0 software (Arizona State University, Arizona, USA) [29], and a NJ tree was constructed based on the ITS2 sequences, with 1000 bootstrap replicates. The ITS2 species determination power was explored by the BLAST1 method, which is based on the best hit of the query sequence and an E-value for the match of less than a cutoff value, as described previously [18]. The secondary structures of the ITS2 sequences were predicted according to an online ITS2 database [30].
Results
DNA extraction and the efficiency of PCR amplification
The genomic DNA extracted from the root samples were extensively degraded and produced faint, diffuse bands upon DNA gel electrophoresis. The success rate for the ITS2 PCR amplification from all the samples was at 100 % (Additional file 1). Moreover, high-quality trace files were obtained for the sequenced ITS2 regions.
Sequence and inter−/intra− specific variation analysis
The intraspecific variation among the 43 samples of Glehniae Radix collected from different localities was not detected (including two partial sequences, GU395183 and AY146915). The ITS2 sequences generated in this study were identical to those from GenBank. We also found that the ITS2 regions of the commercial Glehniae Radix and the original plants belonged to the same haplotype. The length of these sequences was 228 bp after intraspecific alignment, and the GC content was 58.6 %. The sequence possessed a poly(A) structure at position 94–98 bp.
The Glehniae Radix ITS2 DNA sequences diverged considerably from those of its adulterants, which varied from 202 to 266 bp in length. The average GC content was 56.0 % in all taxa. The length of the ITS2 sequences was 275 bp after interspecific alignment, with 189 bp variable sites (68.7 %). The maximum interspecies variation was 0.987 (belonging to S. tatarinowii), whereas the minimum interspecies divergence was 0.248 (belonging to S. gracilis). The average interspecific distance was 0.468, which was greater than the Glehniae Radix intraspecific distance of 0.000 (Table 2). The genetic relationships between Glehniae Radix and its adulterants within the same family (Umbelliferae), S. gracilis, C. violaceum, and C. smyrnioides, were closer than between Glehniae Radix and other adulterants.
Discrimination power analysis
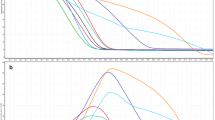
The reliability of the species identification was calculated by the BLAST1, the NJ tree technique and ITS2 sequence secondary structure determination. The ITS2 region exhibited the highest identification efficiency (100 %). Figure 1 shows the phylogenetic tree from 105 ITS2 sequences represented Glehniae Radix and its seven adulterant species. In the cluster dendrogram, G. littoralis samples from different locations were monophyletic and clustered on the same branch. Furthermore, the adulterants of different species formed distinct, nonoverlapping clades. Thus, the NJ tree of the ITS2 sequences correctly placed Glehniae Radix and its adulterants with high statistical support (99 %). Figure 2 indicates that the predicted secondary structures of the ITS2 sequences of Glehniae Radix and its adulterants each had unique molecular morphological characteristics with the stem loop size, number, position and spiral angles of the four helices.
The comparison of ITS2 secondary structures in Glehniae Radix and its adulterants. a Adenophora polyantha HQ704524; b Chuanminshen violaceum GQ434691; c Adenophora tetraphylla AY548194; d Silene tatarinowii FJ384025; e Sphallerocarpus gracilis AF073678; f Adenophora stricta AF090713; G G. littoralis FJ593179; h Changium smyrnioides HQ185237
Discussion
Previous studies have successfully used 5S rRNA spacer domains to distinguish the original G. littoralis plant from its related medicinal species [10, 13] These studies extracted genomic DNA samples from leaves, but did not mention commercial Glehniae Radix samples. A study [31] explored the utility of ITS2 sequences for barcoding Glehniae Radix, their results displayed the similar trend that Glehnia Radix and its three adulterants could be identified by the ITS2 region. But the study included only four Glehniae Radix sequences and six sequences derived from three adulterant species. In this study, we used 46 and 59 ITS2 sequences from Glehniae Radix and its seven adulterants, respectively.
The processing procedures of Glehniae Radix are as following: remove the fibrous roots, stems and residual impurities, wash, slightly dry, rear blanched in boiling water, remove the skin, cut and dry. After processing, genomic DNA extracted from the roots of samples was usually severely degraded or contaminated by micro-organisms [32, 33]. Moreover, it was difficult to obtain high-quality DNA from samples enriched in polysaccharide polyphenols, fibre or other storage materials [17, 24]. In this study, we added 10 % PVP-30 powder when grinding the sample in liquid nitrogen to remove contaminants (polyphenols and polysaccharides). Subsequently, we added nuclear separation liquid to the plant powder 2–3 times, and extended water bath time to ensure DNA could be fully released into the extraction buffer. Additionally, root herbs often contain soil fungi; therefore, to avoid the influence of fungal contamination, one should clean the herb’s surface and isolate the interior material during the sampling procedure. We successfully extracted genomic DNA from 59 various herb samples, including 38 commercial crude drugs and 16 dried root samples.
Because of the DNA degradation of herbal medicines, DNA barcodes that are favourable for some plant species such as rbcL and matK cannot be used to identify commercial herbs [34]. In our study, the DNA isolated from the samples was severely degraded; however, 59 different versions of the ITS2 barcodes were readily retrievable due to the short length of the ITS2 in these samples (202–266 bp). Shorter fragments are easier to amplify from herbarium DNA [35], and the length of the ITS2 region might be sufficient to allow amplification without high-quality DNA [19, 24, 32]. Although multiple sequences were detected from a single individual due to its multicopy genes [36], intragenomic ITS2 variation typically occurred at only a very few, extremely variable, positions; thus, the ITS2 region can be treated as a single gene [37–39]. In our study, 46 sequences of the Glehniae Radix ITS2 barcode were obtained and were identical to each other. Xin et al. [40] also recognized that the application of the ITS2 barcode was not affected by the presence of multiple copies in Goji.
A successful barcode sequence requires both low intraspecific variation and high interspecific divergence. The 46 samples of G. littoralis analysed in this study, which were from original plants and commercial crude drugs, were representatives selected from extensive distribution areas, and their characteristics varied. The ITS2 sequences of G. littoralis within a given sample pool all belonged to the same haplotype, supporting Mizukami et al.’s assertion [41] that the genetic diversity among geographical strains of G. littoralis is narrow. The ITS2 intraspecific genetic distances in the medicinal Panax species were low [39]. Moreover, ITS2 displayed considerable interspecific divergence between Glehniae Radix and its adulterants. Thus, the ITS2 sequence as a barcode shows strong stability within species and high variability between species. Similar results have been found in Flos Lonicerae Japonicae (Jinyinhua) [23]. ITS2 sequences could be suitable for Glehniae Radix identification due to high conservation of the ITS2 regions derived from commercial crude drugs and original plant samples.
An NJ tree was constructed and compared the molecular morphological features of the ITS2 secondary sequences of Glehniae Radix and its adulterants. In the cluster dendrogram, each of the medicinal species was unambiguously distinguishable from each of the others. Furthermore, the secondary structure of ITS2 is considered a molecular morphological characteristic [30, 37]. This study demonstrated that ITS2 has a powerful identification capability, as the ITS2 sequences readily distinguished Glehniae Radix from its adulterants. DNA barcode discrimination technology has been applied to identify commercial plant products in teas [42] and Hypericum species [43], among others [26, 44].
Conclusion
DNA barcoding based on ITS2 distinguished commercial processed Glehniae Radix from common herbal adulterants.
Abbreviations
- ITS2:
-
The second internal transcribed spacer
- NJ:
-
neighbor-joining
- ITS:
-
internal transcribed spacer
- K2P:
-
kimura 2–parameter
- CM:
-
Chinese medicine
- BOL:
-
the Barcode of Life Plant Working Group
- CBOL:
-
The China Plant BOL Group
- PVP:
-
Polyvinyl pyrrolidone
References
The State Pharmacopoeia Commission of the PRC. Pharmacopoeia of the People’s Republic of China. Beijing: Chinese Medical Science and Technology; 2010.
Miyazawa M, Kurose K, Itoh A, Hiraoka N. Comparison of the Essential Oils of Glehnia littoralis from Northern and Southern Japan. J Agr Food Chem. 2001;49:5433–6.
Li J, Cheung AWH, Bi CWC, Duan R, Zheng KYZ, Huang W, Chen JJ, Dong TTX, Tsim KWK. Quality evaluation of Radix Glehniae (Glehnia littoralis) by HPLC-DAD: chromatographic fingerprinting and quantitative analysis of the herbs from different regions of China. Asian J Tradit Med. 2010;5:40–8.
Ksouri R, Ksouri WM, Jallali I, Debez A, Magné C, Hiroko I, Abdelly C. Medicinal halophytes: potent source of health promoting biomolecules with medical, nutraceutical and food applications. Crit Rev Biotechnol. 2012;32:289–326.
Matsuura H, Saxena G, Farmer SW, Hancock RE, Towers GH. Antibacterial and antifungal polyine compounds from Glehnia littoralis. Planta Med. 1996;62:256–9.
Huang GJ, Deng JS, Liao JC, Hou WC, Wang SY, Sung PJ, Kuo YH. Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory activity of imperatorin from Glehnia littoralis. J Agr Food Chem. 2012;60:1673–81.
Fu LK. China plant red data book, Beijing: Science; 1992.
Bi CL. Identification of Radix Glehniae and Its Counterfeit. Shi Zhen Guo Yi Guo Yao. 2003;14:154.
Tu P, Leng Q, Xu G, Xu L. Identification of crude drug, laiyangshen. Zhong Yao Cai. 1999;22:174–6.
Zhao KJ, Dong TTX, Cui XM, Tu PF, Tsim KWK. Genetic distinction of radix adenophorae from its adulterants by the DNA sequence of 5S-rRNA spacer domains. Am J Chinese Med. 2003;31:919–26.
Yuan Z, Tezuca Y, Fan WZ, Kadota S, Li X. Constituents of the underground parts of Glehnia littoralis. Chem Pharm Bull. 2002;50:73–7.
Mizukami H. Amplification and sequence of a 5SrRNA gene spacer region from the Crude Drug “Angelica Root”. Biol Pharm Bull. 1995;18:1299–301.
Chen S, Pang X, Song J, Shi L, Yao H, Han J, Leon C. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 2014;32:1237–44.
Hebert PDN, Ratnasingham S, DeWaard JR. Barcoding animal life: cytochrome coxidase subunit1divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):96–9.
Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–21.
CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–7.
Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25:103–10.
Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, Luo K, Li Y, Li X, Jia X, Lin Y, Leon C. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613.
Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, Chen S. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102.
Gutteridge A, Burns M. The application of DNA molecular approaches for the identification of herbal medicinal products. J Asso Public Analysts. 2013;41:53–66.
Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 2011;108(19641–6):23.
Pang X, Shi L, Song J, Chen X, Chen S. Use of the potential DNA barcode ITS2 to identify herbal materials. J Nat Med. 2013;67:571–5.
Hou D, Song J, Shi L, Ma X, Xin T, Han J, Xiao W, Sun Z, Cheng R, Yao H. Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: a case study on Flos Lonicerae Japonicae. BioMed Res Int. 2013;11:121–7.
Sun Z, Chen S. Identification of cortex herbs using the DNA barcode nrITS2. J Nat Med. 2013;67:296–302.
Gao T, Yao H, Song J, Liu C, Zhu Y, Ma X, Chen S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol. 2010;130:116–21.
Newmaster SG, Grguric M, Shanmughanandhan D, Ramalingam S, Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222.
Flora of China. Flora of China Editorial Committee. 2014. http://www.floraofchina.org. Accessed 1 Sept 2014.
Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430(50–7):30.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Schultz J, Müller T, Achtziger, Seibel PN, Dandekar T, Wolf M. The internal transcribed spacer 2 database-a web server for (not only) low level phylogenetic analyses. Nucleic Acids Res. 2006;34:W704–7.
Li Z, Wang Y, Yuan Z. Identification of Glehniae Radix and its adulterants by ITS2 sequence. Shenyang Yao Ke Da Xue Xue Bao. 2013;30:803–6.
Chiou SJ, Yen JH, Fang CL, Chen HL, Lin TY. Authentication of medicinal herbs using PCR-amplified ITS2 with specific primers. Planta Med. 2007;73:1421–6.
Pirttilä AM, Hirsikorpi M, Kämäräinen T, Jaakola L, Hohtola A. DNA isolation methods for medicinal and aromatic plants. Plant Mol Biol Rep. 2001; 19: 273a-f.
Guo X, Wang X, Su W, Zhang G, Zhou R. DNA barcodes for discriminating the medicinal plant Scutellaria baicalensis (Lamiaceae) and its adulterants. Biol Pharm Bull. 2011;34:1198–203.
Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One. 2012;7:e43808.
Chase MW, Cowan RS, Hollingsworth PM, van den BC, Madriñán S, Peterson G, Seberg O, Jørgsensen T, Cameron KM, Carine M; Pederson N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough T, Kelly L, Wilkinson MJ. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007; 56:295–9.
Coleman AW. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007;35:3322–9.
Song J, Shi L, Li D, Sun Y, Niu Y, Chen Z, Luo H, Pang X, Sun Z, Liu C, Lv A, Deng Y, Larson-Rabin Z, Wilkinson M, Chen S. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS One. 2012;7:e43971.
Chen XC, Liao BS, Song JY, Pang XH, Han JP, Chen SL. A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene. 2013;530:39–43.
Xin T, Yao H, Gao H, Zhou X, Ma X, Xu C, Chen J, Han J, Pang X, Xu R, Song J, Chen S. Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res Int. 2013;54:1699–704.
Mizukami H, Ohbayashi K, Umetsu K, Hiraoka N. Restriction fragment length polymorphisms of medicinal plants and crude drugs. II. Analysis of Glehnia littoralis of different geographical origin. Biol Pharm Bull. 1993;16:611–2.
Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S, Little DP. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep. 2011. doi:10.1038/srep00042.
Howard C, Socratous E, Williams S, Graham E, Fowler MR, Scott NW, Bremner PD, Slater A. Plant ID–DNA-based identification of multiple medicinal plants in complex mixtures. Chin Med. 2012;7:18.
Kool A, Boer HJ, Kruger A, Rydberg A, Abbad A, Bjork L. Martin Gary. Molecular identification of commercialized medicinal plants in southern Morocco. PLoS One. 2012;7:e39459.
Authors’ contributions
TG and XZ designed the study. TG, YZ, XL, and DH performed the experiment. XZ and TG analyzed the data and wrote the manuscript. TG, XZ, and DH revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Startup Foundation for Advanced Talents of Jiangsu University of Science and Technology (No. 635211204), National Natural Science Foundation of China(No. U1404829), the Scientific and Technical Development Project of Qingdao (No. 14-2-4-89-jch), the Natural Science Foundation of Shandong Province, China (No. ZR2013HL021) and a Project of Shandong Province Higher Educational Science and Technology Program, China (No. J14LE13).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional file
13020_2015_71_MOESM1_ESM.jpg
Additional file 1. Results from PCR amplification of the ITS2 regions of G. littoralis M: marker, 1, 18 and 33: negative control (CK), 2–17, 19–32, 33–46: G. littoralis.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhu, X., Zhang, Y., Liu, X. et al. Authentication of commercial processed Glehniae Radix (Beishashen) by DNA barcodes. Chin Med 10, 35 (2015). https://doi.org/10.1186/s13020-015-0071-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-015-0071-8