Abstract
Background
Upland cotton is one of the utmost significant strategic fiber crops, and play a vital role in the global textile industry.
Methods and results
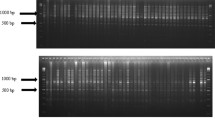
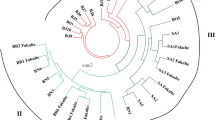
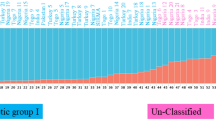
A total of 128 genotypes comprised Gossypium hirsutum L, Gossypium barbadense L., and pure lines were used to examine genetic diversity using iPBS-retrotransposon markers system. Eleven highly polymorphic primers yielded 287 bands and 99.65% polymorphism was recorded. The mean polymorphism information content was estimated at 0.297 and the average diversity indices for the effective number of alleles, Shannon’s information index, and overall gene diversity were 1.481, 0.443, and 0.265, respectively. The analysis of molecular variance (AMOVA) revealed that 69% of the genetic variation was within the population. A model-based STRUCTURE algorithm divided the entire germplasm into four populations and one un-classified population, the genotypes G42 (originating in Egypt) and G128 (originating in the United States), showed the highest genetic distance (0.996) so these genotypes could be suggested for breeding programs as parental lines.
Conclusions
This is the first investigation using an iPBS-retrotransposon marker system to examine the genetic diversity and population structure of upland cotton germplasm. The rich diversity found in upland cotton germplasm could be exploited as a genetic resource when developing breeding programs and could also help with efforts to breed cotton around the world. These findings also show the applicability and effectiveness of iPBS-retrotransposons for the molecular characterization of cotton germplasm.





Similar content being viewed by others
Data Availability
The datasets used for the current study ıs provided in this manuscript.
References
Khan MA, Wahid A, Ahmad M, Tahir MT, Ahmed M, Ahmad S, Hasanuzzaman M (2020) World Cotton Production and Consumption: an overview. In: Ahmad S, Hasanuzzaman M (eds) Cotton Production and uses. Springer, Singapore. https://doi.org/10.1007/978-981-15-1472-2_1
Xiao G, Zhao P, Zhang Y (2019) A pivotal role of hormones in regulating cotton fiber development. Front Plant Sci 10(87). https://doi.org/10.3389/fpls.2019.00087
Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V (2014) Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L). Theor Appl Genet 127(2):283–295. https://doi.org/10.1007/s00122-013-2217-3
Jabran K, Ul-Allah S, Chauhan BS, Bakhsh A (2019) An Introduction to Global Production Trends and Uses, History and Evolution, and Genetic and Biotechnological Improvements in cotton. Hoboken, NJ: wiley online library. https://doi.org/10.1002/9781119385523.ch1
Saleem MA, Qayyum A, Malik W, Amjid MW (2020) Molecular breeding of cotton for Drought stress tolerance. In: Ahmad S, Hasanuzzaman M (eds) Cotton Production and uses. Springer, Singapore. https://doi.org/10.1007/978-981-15-1472-2_1
Basal H, Karademir E, Goren HK, Sezener V, Dogan MN, Gencsoylu I, Erdogan O (2019) Cotton production in turkey and Europe. In: Jabran K, Chauhan BS (eds) Cotton Production. John Wiley & Son Inc., New York, NY, USA, pp 297–321
Hayat K, Bardak A, Parlak D, Ashraf F, Imran HM, Haq HA, Mian MA, Mehmood Z, Akhtar MN (2020) Biotechnology for Cotton Improvement. In: Ahmad S, Hasanuzzaman M (eds) Cotton Production and uses. Springer, Singapore. https://doi.org/10.1007/978-981-15-1472-2_1.
Malik W, Ashraf J, Iqbal MZ, Khan AA, Qayyum A, Abid MA, Noor E, Ahmad MQ, Abbasi GH (2014) Molecular markers and cotton genetic improvement: Current status and future prospects. The Scientific World Journal, 2014, 1–15. https://doi.org/10.1155/2014/607091
Tokel D, Dogan I, Hocaoglu-Ozyigit A, Ozyigit II (2022) Cotton Agriculture in Turkey and Worldwide Economic Impacts of turkish cotton. J Nat Fib 6:1–20
USDA, United States Department of Agriculture Foreign Agricultural Service (2020a), December 2020 Report, Cotton: World Markets and Trade, 1–28. https://downloads.usda.library.cornell.edu/usda-esmis /
Yilmaz I, Akcaoz H, Ozkan B (2005) An analysis of energy use and input costs for cotton production in Turkey. Renewable Energy 30:145–155. https://doi.org/10.1016/j.renene.2004.06.001
Daǧdelen N, Başal H, Yilmaz E, Gürbüz T, Akçay S (2009) Different drip irrigation regimes affect cotton yield, water use efficiency and fiber quality in western Turkey. Agric Water Manage 96(1):111–120. https://doi.org/10.1016/j.agwat.2008.07.003
Rana MK, Bhat KV (2005) RAPD markers for genetic diversity study among indian cotton cultivars. Curr Sci 88(12):1956–1961
Dongre AB, Bhandarkar M, Banerjee S (2007) Genetic diversity in tetraploid and diploid cotton (Gossypium spp.) using ISSR and microsatellite DNA markers. Indian J Biotechnol 6:349–353
Li Z, Wang X, Zhang Y, Zhang G, Wu L, Chi J, Ma Z (2008) Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Front Agric China 2(3):245–252. https://doi.org/10.1007/s11703-008-0063-x
Murtaza N (2006) Cotton genetic diversity study by AFLP markers. Electron J Biotechnol 9(4):1–5. https://doi.org/10.2225/vol9-issue4-fulltext-9
Hocaoglu-ozyigit A, Ucar B, Altay V, Ozyigit II (2020) Genetic diversity and phylogenetic analyses of turkish cotton (Gossypium hirsutum L.) lines using ISSR markers and Chloroplast trnL-F regions. J Nat Fibers 1–14. https://doi.org/10.1080/15440478.2020.1788493
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32(2):261–285. https://doi.org/10.1080/13102818.2017.1400401
Baloch FS, Alsaleh A, de Miera LES, Hatipoğlu R, Çifti V et al (2015) DNA based iPBS-retrotransposon markers for investigating the population structure of pea (Pisum sativum) germplasm from Turkey. Biochem Syst Ecol 61:244–252. https://doi.org/10.1016/j.bse.2015.06.017
Yıldız M, Koçak M, Baloch FS (2015) Genetic bottlenecks in turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment. Genet Mol Res 14:10588–10602. https://doi.org/10.4238/2015.September.8.20
Demirel U, Tındaş İ, Yavuz C, Baloch F, Çalışkan M (2018) Assessing genetic diversity of potato genotypes using inter-PBS retrotransposon marker system. Plant Genetic Resources: Characterization and Utilization 16(2):137–145. https://doi.org/10.1017/S1479262117000041
Yaldız G, Camlica M, Nadeem MA, Nawaz MA, Baloch FS (2018) Genetic diversity assessment in Nicotiana tabacum L. with iPBS-retrotransposons. Turk J Agric For 42:154–164. https://doi.org/10.3906/tar-1708-32
Ali F, Yılmaz A, Nadeem MA, Habyarimana E, Subaşı I et al (2019) Mobile genomic element diversity in world collection of safower (Carthamus tinctorius L.) panel using iPBS-retrotransposon markers. PLoS ONE 14(2):1–19. https://doi.org/10.1371/journal.pone.0211985
Yıldız M, Koçak M, Nadeem MA, Cavagnaro P, Barboza K et al (2019) Genetic diversity analysis in the turkish pepper germplasm using iPBS retrotransposon-based markers. Turk J Agric For 44:1–14. https://doi.org/10.3906/tar-1902-10
Barut M, Nadeem MA, Karaköy T, Baloch FS (2020) DNA fngerprinting and genetic diversity analysis of world quinoa germplasm using iPBS-retrotransposon marker system. Turk J Agric For 44:479–491. https://doi.org/10.3906/tar-2001-10
Shimira F, Boyaci HF, Yeter Ç, Nadeem MA, Baloch FS, Taskin H (2021) Exploring the genetic diversity and population structure of scarlet eggplant germplasm from Rwanda through iPBS retrotransposon markers. Mol Biol Rep. https://doi.org/10.1007/s11033-021-06626-0
Baloch FS, Guizado SJ, Altaf MT, Yüce I, Çilesiz Y, Bedir M, Nadeem MA, Hatipoglu R, Gómez JC (2022) Applicability of inter-primer binding site iPBS-retrotransposon marker system for the assessment of genetic diversity and population structure of peruvian rosewood (Aniba rosaeodora Ducke) germplasm. Mol Biol Rep 49(4):2553–2564
Kalendar R, Antonius K, Smýkal P, Schulman HA (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430. https://doi.org/10.1007/s00122-010-1398-2
Milovanov A, Zvyagin A, Daniyarov A et al (2019) Genetic analysis of the grapevine genotypes of the russian Vitis ampelographic collection using iPBS markers. Genetica 147:91–101. https://doi.org/10.1007/s10709-019-00055-5
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Baloch, F. S., Guizado, S. J. V., Altaf, M. T., Yüce, I., Çilesiz, Y., Bedir, M.,… Gómez, J. C. C. (2022). Applicability of inter-primer binding site iPBS-retrotransposon marker system for the assessment of genetic diversity and population structure of Peruvian rosewood (Aniba rosaeodora Ducke) germplasm. Molecular Biology Reports, 49(4),2553–2564
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. Molecular Biology and Biotechnology Centre—University of Alberta, Edmonton
Roldán-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp). Mol Breed 6(2):125–134. https://doi.org/10.1023/A:1009680614564
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Adawy SS, Hussein EHA, El-itriby HA (2006) Molecular characterization and genetic relationships among cotton genotypes 2- AFLP analysis. Arab J Biotechnol 9(3):477–492
Esmail RM, Zhang JF, Adel-Hamid AM (2008) Genetic diversity in Elite Cotton Germplasm Lines using field performance and rapd markers. World J Agricultural Sci 4(3):369–375
Liu Z, Zhang Y, Zhang S, Deng K, Dong S, Ren Z (2011) Genetic diversity analysis of various red spider mite- resistant upland cotton cultivars based on RAPD. Afr J Biotechnol 10(18):3515–3520. https://doi.org/10.5897/AJB10.832
Noormohammadi Z, Shojaei-jesvaghani F, Sheidai M, Farahani F, Alishah O (2011) Inter simple sequence repeats (ISSR) and random amplified polymorphic DNA (RAPD) analyses of genetic diversity in Mehr cotton cultivar and its crossing progenies. Afr J Biotechnol 10(56):11839–11847. https://doi.org/10.5897/AJB11.1377
Noormohammadi Z, Farahani YH, Sheidai M, Ghasemzadeh-Baraki S, Alishah O (2013a) Genetic diversity analysis in Opal cotton hybrids based on SSR, ISSR, and RAPD markers. Genet Mol Res 12(1):256–269. https://doi.org/10.4238/2013.January.30.12
Zhang Y, Wang XF, Li ZK, Zhang GY, Ma ZY (2011) Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet Mol Res 10(3):1462–1470. https://doi.org/10.4238/vol10-3gmr1277
De Magalhães Bertini CHC, Schuster I, Sediyama T, de Barros EG, Moreira MA (2006) Characterization and genetic diversity analysis of cotton cultivars using microsatellites. Genet Mol Biology 29(2):321–329. https://doi.org/10.1590/S1415-47572006000200021
Chen G, Du, Xiong-ming D (2006) Genetic diversity of source germplasm of Upland Cotton in China as determined by SSR marker analysis. Acta Genetica Sinica 33(8):733–745. https://doi.org/10.1016/S0379-4172(06)60106-6
Khan AI, Fu Y, Khan IA (2009) Genetic diversity of pakistani cotton cultivars as revealed by simple sequence repeat markers. Commun Biometry Crop Sci 4(1):21–30
Campbell BT, Williams VE, Park W (2009) Using molecular markers and w eld performance data to characterize the Pee Dee cotton germplasm resources. Euphytica 169:285–301. https://doi.org/10.1007/s10681-009-9917-4
Tu JL, Zhang MJ, Wang XQ, Zhang XL, Lin ZX (2014) Genetic dissection of upland cotton (Gossypium hirsutum) cultivars developed in Hubei Province by mapped SSRs. Genet Mol Res 13(1):782–790. https://doi.org/10.4238/2014.January.31.4
Ge H, Liu Y, Jiang M, Zhang J, Hana H et al (2013) Analysis of genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat markers. Sci Hortic 162:71–75. https://doi.org/10.1016/j.scienta.2013.08.004
De Menezes IPP, Hoffmann LV, Barroso APV (2015) Genetic characterization of cotton landraces found in the Paraíba and Rio Grande do norte states. Crop Breed Appl Biotechnol 15:26–32. https://doi.org/10.1590/1984-70332015v15n1a4
Noormohammadi Z, Rahnama A, Sheidai M (2013b) EST-SSR and SSR analyses of genetic diversity in diploid cotton genotypes from Iran. Nucleus 56(3):171–178. https://doi.org/10.1007/s13237-013-0094-4
Yildiz E, Sümbül A, Yaman M, Nadeem MA, Say A, Baloch FS, Popescu GC (2022) Assessing the genetic diversity in hawthorn (Crataegus spp.) genotypes using morphological, phytochemical and molecular markers. Genetic Resources and Crop Evolution, pp 1–12
Baloch FS, Guizado SJV, Altaf MT, Yüce I, Çilesiz Y, Bedir M, Gómez JCC (2022) Applicability of inter-primer binding site iPBS-retrotransposon marker system for the assessment of genetic diversity and population structure of peruvian rosewood (Aniba rosaeodora Ducke) germplasm. Mol Biol Rep 49(4):2553–2564
Alsaleh A, Bektas H, Baloch FS, Nadeem MA, Özkan H (2022) Turkish durum wheat conserved ex-situ and in situ unveils a new hotspot of unexplored genetic diversity. Crop Sci 62(3):1200–1212
Bouchet S, Pot D, Deu M, Rami JF, Billot C, Perrier X et al (2012) Genetic structure, linkage disequilibrium and signature of selection in sorghum: lessons from physically anchored DArT markers. PLoS ONE 7:3
Nadeem MA (2021) Deciphering the genetic diversity and population structure of turkish bread wheat germplasm using iPBS-retrotransposons markers. Mol Biol Rep 48(10):6739–6748
Acknowledgements
We would like to thank the Nazilli Cotton Research Institute and Prof. Dr. Mefhar Gültekin TEMİZ from the Faculty of Agriculture, University of Dicle (Diyarbakir, Turkey) for their assistance in supplying the cotton genetic materials used in this study.
Funding
Authors are grateful to Dicle University - Scientific Research Projects Coordinating Office for financial support (Project No: ZİRAAT.20.004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no conflicts of interests.
Consent to publish
All authors read the manuscript and showed their willingness to publish this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baran, N., Shimira, F., nadeem, M.A. et al. Exploring the genetic diversity and population structure of upland cotton germplasm by iPBS-retrotransposons markers. Mol Biol Rep 50, 4799–4811 (2023). https://doi.org/10.1007/s11033-023-08399-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08399-0