Abstract
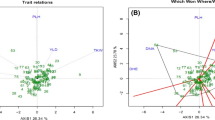
Seventy-one glandless cotton germplasm resources were firstly evaluated genetically by using nine agronomic traits, 33 simple sequence repeat (SSR) primers and ten amplified fragment length polymorphism (AFLP) primer combinations. Principal component analysis (PCA) of the agronomic traits showed that the first six principal components (PCs) explained a total of 86.352% of the phenotypic variation. A total of 329 alleles were amplified for 33 SSR primers, and 232 polymorphic bands in a total of 389 bands were obtained by using ten AFLP primer combinations. The average polymorphic information content (PIC) value was 0.80 and 0.18 for SSR primers and AFLP primer combinations, respectively. The DIST (average taxonomic distance) and DICE (Nei and Li’s pairwise distance) coefficients ranged from 0.373 to 3.164 and 0.786 to 0.948, respectively, for agronomic traits and SSR&AFLP data based on UPGMA analysis. This suggested that there was a higher diversity in the evaluated population for both agronomic traits and molecular markers. The Mantel’s test showed that the correlation between the dendrograms based on agronomic traits and SSR&AFLP data was non-significant.
Similar content being viewed by others
References
Abdalla A M, Reddy OUK, El-Zik K M, Pepper A E (2001). Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet, 102: 222–229
Afifi A, Bary A A, Kamel S A, Heikal I (1966). Bahtim 110, a new strain of Egyptian cotton free from gossypol. Cot Grow Rev, 43: 112–120
Beyene Y, Botha A M, Myburg A A (2005). A comparative study of molecular and morphological methods of describing genetic relationships in traditional Ethiopian highland maize. Afri J Biot, 4(7): 586–595
Bottger G T, Sheehan E T, Lukefahr M J (1964). Relation of gossypol content of cotton plant to insect resistance. J Economic Entomo, 57(2): 283–285
Cai Y F, Zhang H, Zeng Y, Mo J C, Bao J K, Miao C, Bai J, Yan F, Chen F (2004). An optimized gossypol high-performance liquid chromatography assay and its application in evaluation of different gland genotypes of cotton. J Biosci, 29: 67–71
De Riek J, Calsyn E, Everaert I, van bockstaele E, De Loose M (2001). AFLP based alternatives for the assessment of distinctness uniformity and stability of sugar beet varieties. Theor Appl Genet, 103: 1254–1265
James Rohlf F (2000). NTSYSpc Numerical Taxonomy and Multivariate Analysis System Version 2.1 User Guide. Department of Ecology and Evolution State University of New York Stony Brook, NY, 11794–5245
Lee J A (1962). Genetical studies concerning the distribution of pigment glands in the cotyledons and leaves of upland cotton. Genetics, 47: 131–142
Liu S, Cantrell R G, McCarty J C Jr, Stewart J McD (2000)a. Simple sequence repeat-based assessment of genetic diversity in cotton race stock accessions. Crop Sci, 40: 1459–1469
Liu S, Saha S, Stelly D, Burr B, Cantrell RG (2000)b. Chromosomal assignment of microsatellite loci in cotton. J Hered, 91: 326–332
Mantel N (1967). The detection of disease clustering and a generalized regression approach. Cancer Res, 27: 209–220
McMichael S C (1954). Glandless boll in upland cotton and its use in the study of natural crossing. J Agron, 46: 527–528
McMichael S C (1959). Hopi cotton: a source of cottonseed free of gossypol pigments. J Agron, 51: 630
McMichael S C (1960). Combined effects of the glandless genes gl2 and gl3 on pigment glands in the cotton plant. J Agron, 52: 385–386
Murray J C (1965). A new locus for glanded atem in tetraploid cotton. J Heredity, 56: 42–46
Nei M, Li W H (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci, 76: 5269–5273.
Ni J J, Colowit P M, Mackill D J (2002). Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci, 42: 601–607
Paterson AH, Brubaker C L, Wendel J F (1993). A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep, 11(2): 122–127
Rana M K, Singh V P, Bhat K V (2005). Assessment of genetic diversity in upland cotton (Gossypium hirsutum L.) breeding lines by using amplified fragment length polymorphism (AFLP) markers and morphological characteristics. Genet Res and Crop Evol, 52: 989–997
Reddy A S, Haisler RM, Yu Z H, Kohel R J (1997). AFLP mapping in cotton. Plant and Animal Genome V Conference, 80
Reinisch A R, Dong JM, Brubaker C, Stelly D, Wendel J, Patterson A H (1994). A detailed RFLP map of cotton, Gossypium hirsutum6Gossypium barbadense: Chromosome organization and evolution in a disomic polyploid genome. Genetics, 138: 829–847
Roux J B (1960). The selection of cotton plants without gossypol. C R Acad Agr Fr, 46: 613–622 (in French)
Saha S, Karaca M, Jenkins J N, Zipf A E, Reddy U K, Kantety R V (2003). Simple sequence repeats as useful resources to study transcribed genes of cotton. Euphytica, 130: 355–364
Schondelmaier J, Steinrucken G, Jung C (1996). Integration of AFLP markers into a linkage map of sugar beet (Beta vulgaris L.). Plant Breeding, 115: 231–237
Shappley Z W, Jenkins J N, Meredith W R, McCarty J C Jr (1998). An RFLP linkage map of upland cotton, Gossypium hirsutum L. Theor Appl Genet, 97: 756–761
Tara S C, Bhat K V, Bharadwaj C, Tiwari S P, Chaudhury V K (2006). AFLP analysis of genetic diversity in Indian soybean [Glycine max (L.) Merr.] varieties. Gene Res and Crop Evol, 53: 1069–1079
van Esbroeck G A, Bowman D T (1998). Cotton germplasm diversity and its importance to cultivar development. J Cot Sci, 2: 121–129
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Fijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res, 23(21): 4407–4414
Vroh Bi I, Maquet A, Bandoin J P, Jardin P du, Jacquemin J M, Mergeai G (1999). Breeding for “low gossypol seed and high gossypol plants” in upland cotton. Analysis of tri-species hybrids and backcross progenies using AFLPs and RFLPs. Theor Appl Genet, 99: 1233–1244
Warburton M, Crossa J (2002). Data analysis in the CIMMYT applied biotechnology center for fingerprinting and genetic diversity studies. 2nd ed.www.cimmyt.org/english/docs/manual/protocols/dataAnalysis.pdf
Zhang J F, Lu Y, Adragna H, Hughs E (2005)a. Genetic improvement of new Mexico Acala cotton germplasm and their genetic diversity. Crop Sci, 45: 2363–2373
Zhang J F, Lu Y, Cantrell R G, Hughs E (2005)b. Molecular marker diversity and field performance in commercial cotton cultivars evaluated in the southwestern USA. Crop Sci, 45: 1483–1490
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Li, Z., Wang, X., Zhang, Y. et al. Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Front. Agric. China 2, 245–252 (2008). https://doi.org/10.1007/s11703-008-0063-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11703-008-0063-x