Abstract
Background
Research activities aiming to investigate the genetic diversity are very crucial because they provide information for the breeding and germplasm conservation activities. Wheat is one of the most important cereal crops globally by feeding more than a third of the human population around the world.
Methods and results
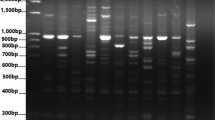
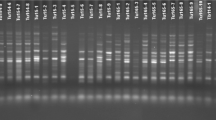
During present investigation, a total of 74 Turkish bread wheat accessions (54 landraces and 20 cultivars) were used as plant material and iPBS-retrotransposons marker system was used for the molecular characterization. 13 polymorphic primers used for molecular characterization resulted a total of 152 bands. Range of calculated diversity indices like polymorphism information content (0.11–0.702), effective numbers of alleles (1.026–1.526), Shannon’s information index (0.101–0.247) and gene diversity (0.098–0.443) confirmed higher genetic variations in studied germplasm. Bread wheat landraces reflected higher genetic variations compared to commercial cultivars. Analysis of molecular variance resulted that higher (98%) genetic variations are present within populations. The model-based structure algorithm separated 74 bread wheat accessions in to two populations. Diversity indices based on structure evaluated population’s revealed population B as a more diverse population. The principal coordinate analysis and neighbor-joining analysis separated 74 bread wheat accessions according to their collection points. Genetic distance for 74 Turkish bread wheat accessions explored Bingol and Asure accessions as genetically diverse that can be used as parents for breeding activities.
Conclusions
The extensive diversity of bread wheat in Turkish germplasm might be used as genetic resource for the exhaustive wheat breeding program. For instance, accessions Bingol and Asure were found genetically diverse and can be used as parents for future breeding activities.





Similar content being viewed by others
Data availability
All data needed to conduct this study is provided within the manuscript.
References
Khush GS, Lee S, Cho JI, Jeon JS (2012) Biofortification of crops for reducing malnutrition. Plant Biotechnol Rep 6:195–202
Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C et al (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–818
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. PNAS 108:20260–20264
Bhatta M, Eskridge KM, Rose DJ, Santra DK, Baenziger PS, Regassa T et al (2017) Seeding rate, genotype, and topdressed nitrogen effects on yield and agronomic characteristics of winter wheat. Crop Sci 57:951–963
Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, Wulff BB, Steuernagel B, Mayer KF, Olsen OA (2014) International wheat genome sequencing consortium ancient hybridizations among the ancestral genomes of bread wheat. Science 345(6194):1250092. https://doi.org/10.1126/science.1250092
Feldman M, Lupton FG, Miller TE (1995) Wheat Triticum spp. (Gramineae-Triticinae). In: Smartt J, Simmonds NW (eds) Evolution of crop plants. Longman Scientific and Technical, New York, pp 184–192
Huang X, Wang L, Xu M, Röder M (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106(5):858–865
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316(5833):1862–1866
Food and Agriculture Organization (2021) FAO statistical yearbook. Available at http://www.fao.org/docrep/017/i3138e/i3138e. Accessed 17 June 2021
Karagöz P, Özkan M (2014) Ethanol production from wheat straw by Saccharomyces cerevisiae and Scheffersomyces stipitis co-culture in batch and continuous system. Biores Technol 1(158):286–293
Morgounov AI, Keser M, Kan M, Kucukcongar M, Ozdemir F, Gummadov N, Muminjanov H, Zuev E, Qualset C (2016) Wheat landraces currently grown in Turkey: distribution, diversity, and use. Crop Science Society of America, Fitchburg
Nadeem MA, Gündoğdu M, Ercişli S, Karaköy T, Saracoğlu O, Habyarimana E, Lin X, Hatipoğlu R, Nawaz MA, Sameeullah M, Ahmad F, Baloch FS et al (2020) Uncovering phenotypic diversity and DArTseq marker loci associated with antioxidant activity in common bean. Genes. https://doi.org/10.3390/j.genes.2020.11010036
Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J et al (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol 24:1506–1517
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Baloch FS et al (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32:261–285
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103–107. https://doi.org/10.1016/0168-9525(89)90039-5
Alzohairy AM, Gyulai G, Ramadan MF, Edris S, Sabir JS, Jansen RK, Eissa HF, Bahieldin A et al (2014) Retrotransposon-based molecular markers for assessment of genomic diversity. Funct Plant Biol 41:781–789
Kalendar R, Antonius K, Smýkal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430. https://doi.org/10.1007/s00122-010-1398-2
Karık Ü, Nadeem MA, Habyarimana E, Ercişli S, Yildiz M, Yılmaz A, Yang SH, Chung G, Baloch FS et al (2019) Exploring the genetic diversity and population structure of Turkish laurel germplasm by the iPBS-retrotransposon marker system. Agronomy. https://doi.org/10.3390/agronomy9100647
Baloch FS, Alsaleh A, de Miera LE, Hatipoğlu R, Çiftçi V, Karaköy T, Yıldız M, Özkan H et al (2015) DNA based iPBS-retrotransposon markers for investigating the population structure of pea (Pisum sativum) germplasm from Turkey. Biochem Syst Ecol 61:244–252
Nemli S, Kianoosh T, Tanyolac MB (2015) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based inter primer binding sites (iPBSs) markers. Turk J Agric For 39:940–948
Holasou HA, Rahmati F, Rahmani F, Imani M, Talebzadeh Z (2019) Elucidate genetic diversity and population structure of bread wheat (Triticum aestivum L.) cultivars using IRAP and REMAP markers. JCSB 22:139–151
Khadka K, Torkamaneh D, Kaviani M, Belzile F, Raizada MN, Navabi A et al (2020) Population structure of Nepali spring wheat (Triticum aestivum L.) germplasm. BMC Plant Biol. https://doi.org/10.1186/s12870-020-02722-8
Yadav MK, Chand P (2018) Assessment of genetic diversity among twenty Indian wheat (Triticum aestivum L.) cultivars using simple sequence repeat (SSR) markers. Int J Curr Microbiol App Sci 7:1708–1717
Doyle JJ, Doyle JL (1990) Isolation ofplant DNA from fresh tissue. Focus 12:39–40
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinforma
Roldán-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6(2):125–134. https://doi.org/10.1023/A:1009680614564
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Francis RM (2017) pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 2017:27–32
Demirel F (2020) Genetic diversity of Emmer wheats using iPBS markers. Avrupa Bilim ve Teknoloji Dergisi. https://doi.org/10.31590/ejosat.814537
Queen RA, Gribbon BM, James C, Jack P, Flavell AJ et al (2004) Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol Genet Genom 271(1):91–97
Nazarzadeh Z, Onsori H, Akrami S (2020) Genetic diversity of bread wheat (Triticum aestivum L.) genotypes using RAPD and ISSR molecular markers. J Genet Resour 6:69–76
Kumar P, Sharma V, Sanger RS, Kumar P, Yadav MK et al (2020) Analysis of molecular variation among diverse background wheat (Triticum aestivum L.) genotypes with the help of ISSR markers. IJCS 8:271–276
Alshehri MA, Alzahrani O, Aziza AT, Alasmari A, Ibrahim S, Bahattab O, Osman G, Alshamari AK, Alduaydi SA et al (2020) Correlation and genetic analyses of different characteristics in Saudi Arabian wheat reveal correlation networks and several trait-associated markers. J Anim Plant Sci 30(6):1486–1497
Çifçi EA, Yağdi K (2012) Study of genetic diversity in wheat (Triticum aestıvum) varities using random amplified polymorphic DNA (RAPD) analysis. Turk J Field Crop 17(1):91–95
El-Sherbeny GA, Omara MK, Farrage AA, Khaled AG (2020) Associations between ISSR markers and quantitative traits in bread wheat genotypes. Asian J Biol Sci 2(1):1–8
Al-Tamimi AJ, Al-Janabi AS (2019) Genetic diversity among bread wheat genotypes using RAPD and SSR markers. SABRAO J Breed Genet 51:325–339
Carvalho A, Lima-Brito J, Maçãs B, Guedes-Pinto H (2009) Genetic diversity and variation among botanical varieties of old Portuguese wheat cultivars revealed by ISSR assays. Biochem Genet 47:276–294
Arystanbekkyzy M, Nadeem MA, Aktas H, Yeken MZ, Zencirci N, Nawaz MA, Ali F, Haider MS, Tunc K, Chung G, Baloch FS et al (2019) Phylogenetic and taxonomic relationship of turkish wild and cultivated emmer (Triticum turgidum ssp. dicoccoides) revealed by iPBSretrotransposons markers. IJAB 21:155–163
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z, Moradi Z et al (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30:1075–1081
Shoaib A, Arabi MI (2006) Genetic diversity among Syrian cultivated and landraces wheat revealed by AFLP markers. Genet Resour Crop Evol 53:901–906
Mourad AM, Belamkar V, Baenziger PS (2020) Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genom. https://doi.org/10.1186/s12864-020-06835-0
Darvishzadeh R, Bernousi I (2012) Molecular similarity relationships among Iranian bread wheat cultivars and breeding lines using ISSR markers. Not Bot Horti Agrobot Cluj Napoca 40:254–260
Bouchet S, Pot D, Deu M, Rami JF, Billot C, Perrier X, Rivallan R, Gardes L, Xia L, Wenzl P, Kilian A et al (2012) Genetic structure, linkage disequilibrium and signature of selection in sorghum: lessons from physically anchored DArT markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0033470
Newell MA, Cook D, Hofmann H, Jannink JL (2013) An algorithm for deciding the number of clusters and validation using simulated data with application to exploring crop population structure. Ann Appl Stat 7:1898–1916
Acknowledgements
The author is very grateful to Assoc. Prof. Faheem Shehzad Baloch from Faculty of Agricultural Sciences and Technologies, Sivas University of Science and Technology for providing plant material and supporting during whole experiment and reviewing the manuscript.
Funding
No funding was received to conduct this study.
Author information
Authors and Affiliations
Contributions
MAN conceptualized and established the methodology for this study, also performed molecular characterization of bread wheat germplasm, statistical analysis, and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author confirm that this article content has no conflict of interest.
Consent to publish
Author read the manuscript and showed his willingness to publish this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nadeem, M.A. Deciphering the genetic diversity and population structure of Turkish bread wheat germplasm using iPBS-retrotransposons markers. Mol Biol Rep 48, 6739–6748 (2021). https://doi.org/10.1007/s11033-021-06670-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06670-w