Abstract
Among different types of potato cytoplasmic genomes, some are associated with male sterility or affect agronomic traits. The goal of this study was to analyze types of chloroplast and mitochondrial genomes of selected potato relatives originating from collection of the Institute of Plant Industry, Saint Petersburg, Russia, and preserved in Poland. Using chloroplast and mitochondrial markers the cytoplasm types were determined for 401 genotypes belonging to 43 seed accessions of 28 Solanum species. Among characterized genotypes, 201 (50.1%), 156 (38.9%) and 44 (11%) had cytoplasm types W, D, M, respectively. No accessions with the T, P or A cytoplasm were found. Within cytoplasm W, genotypes with the subtypes: W/α and W/β were identified, but not with W/γ. In S. famatinae, we detected unusual product of the T marker with 65 bp insertion earlier seen exclusively in S. vernei. Among the genotypes of S. leptophyes, two profiles of the ALM_4/ALM_5 marker were observed. S. famatinae and S. vernei come from Argentina, provinces Catamarca and Tucumán. Possibly the insertion in marker T occurred independently in two species, or the accessions were misidentified. Segregation of the ALM_4/ALM_5 marker within S. leptophyes indicates that potato seed accessions are heterogeneous not only due to nuclear DNA polymorphisms but have diversified cytoplasm, too. Our findings are important for exploitation of the tested material in potato breeding. Male-fertile cytoplasm types give a chance of avoiding fertility problems and widening the range of crosses in future generations of breeding materials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato, Solanum tuberosum ssp. tuberosum is an autotetraploid crop, vegetatively propagated with high heterozygosity and strong inbreeding depression. It is characterized by different types of cytoplasmic genomes, among which a sterilizing cytoplasm of S. tuberosum invasively dominates in cultivated potato [1]. This type of cytoplasm is associated with nuclear-cytoplasmic male sterility, which invoked restrictions in sexual hybridization of potato. Today, there is a need of continuous supply of new genetic diversity of potato, because the potato breeding is facing new challenges. Characterization and selection of parents in relation to the cytoplasmic type is the key to the successful breeding programs. Potato relatives are good sources of different types of cytoplasm and due to adaptation to various, often extreme environmental conditions, they show greater tolerance to various environmental stresses and are resistant to many pathogens [2]. Wild potato species are originated from North and South America, mainly from Peru, Mexico, Bolivia, Argentina, Venezuela, Colombia and Ecuador [3].The taxonomic classification of wild potato species is complex and is the subject to constant verification. Wild and cultivated potato species belong to the genus Solanum, section Petota. The Petota section brings together 228 species, grouped in 21 taxonomic series [4]. This number was reduced to 203 [5], then 188 [6], currently it is assumed to be 122 [7].
Chloroplast and mitochondrial DNA are maternally inherited in most of higher plants [8]. In potato, based on the molecular markers specific for mitochondrial (mtDNA) and chloroplast (cpDNA) DNA, five types of mtDNA (α, β, γ, δ, ε) and five types of cpDNA (W, C, T, A, S) were distinguished [9]. It was noticed that a given type of mtDNA is linked with a specific type of cpDNA. Based on that, six types of cytoplasmic DNA: T/β, W/α, W/γ, W/δ, A/ε and S/ε were distinguished [9]. Hosaka and Sanetomo [10] have developed a simpler method of identifying cytoplasmic DNA. Based on six markers specific for mt- and cpDNA, they distinguished six types of cytoplasmic DNA: W (W /α, W/β, W/γ), T (T/β), D (W/α), A (A/ε), P (S/ε) and M (C/ε).
Mutual interactions of nucleus and cytoplasm (plastids/mitochondria) affect many agronomic traits in crop plants. The best known trait controlled by the nuclear and mitochondrial genomes is cytoplasmic male sterility [11]. This trait is manifested in different ways depending on the type of cytoplasm. Type T (T/β) causes cytoplasmic male sterility that manifests as lack of pollen or poor pollen shedding, deformation of anthers or pollen grains. Potato varieties, characterized by the type of cytoplasm W (W/γ), derived from S. stoloniferum, produce small amounts of pollen grains that do not separate in the process of microsporogenesis, forming characteristic tetrads (so called tetrad sterility) [12, 13]. Forms of type D (W/α), derived from S. demissum, produce the right amount, morphologically unchanged pollen, however, unable to fertile with S. tuberosum [13]. Type T (T/β) dominates among cultivated potato varieties, which is a consequence of using these forms as seed parents, because of pollen sterility. It was also noticed that offspring of mothers with T cytoplasm (S. tuberosum ssp. tuberosum) had higher tuber yield, higher tuber number, and earlier vine maturity in comparison with that of A cytoplasm type (S. tuberosum ssp. andigena) [14,15,16,17,18,19].
Potato species and valuable interspecific Solanum hybrids are maintained in many genebank collections worldwide [7]. Chimote and collaborators [20] analyzed variation of chloroplast and mitochondrial genomes in potato varieties and advanced hybrids using PCR markers specific for chloroplast [1] and mitochondrial DNA. The cytoplasm types of Japanese potato collection were determined with type specific DNA markers [10]. The same markers were used for cytoplasmic characterization of European potato cultivars and breeding clones preserved at Max-Planck Institute for Plant Breeding Research [17] and International Potato Center (CIP) potato breeding germplasm [21]. They all proved that sterilizing T-type cytoplasm of S. tuberosum is predominant in analyzed collections and they observed an increasing number of forms with sterilizing D and W/γ cytoplasm types. For these reasons, identification of cytoplasm types and introduction of the fertile ones into new cultivars is crucial for maintaining biodiversity of potato. Potato somatic hybrids are also a valuable source of cytoplasmic diversity. Not only segregation of cytoplasm of parental forms [22] but also changes in cp- and mtDNA have been observed among somatic hybrids [23, 24].
A fraction of wild and cultivated potato species from historical, unique collection of the Institute of Plant Industry (VIR) (Saint Petersburg, Russia) is preserved in Poland, in Plant Breeding and Acclimatization Institute—National Research Institute (IHAR-PIB) [25]. Bukasov, Voronov and Juzepczuk described 30 wild and 18 cultivated species collected in Central and South America [7, 25]. These species were the beginning of the VIR potato collection [7, 25]. True seeds of 111 accessions of wild and cultivated potato species from VIR collection are maintained at IHAR-PIB Genebank. It is a result of reproducing this collection as a part of the Cornell-Eastern Europe-Mexico Project on Late Blight Control [26].
The goal of this study was to analyze types of chloroplast and mitochondrial genomes of selected wild potato species originating from VIR potato collection. In the present study four markers specific to cpDNA and two markers specific to mtDNA were used to investigate the cytoplasmic genome types according to Hosaka and Sanetomo [10].
Materials and methods
Plant materials
Forty-three accessions of 28 tuber-bearing wild potato species were evaluated using a set of cytoplasmic markers. They are preserved at the Plant Breeding and Acclimatization Institute – National Research Institute (IHAR-PIB) as a part originated from VIR collection (Saint Petersburg, Russia) described earlier by Zoteyeva et al. [25]. These species have been selected in order to search for new sources of resistance to P. infestans. Seeds (30 seeds per accession) were sown in three years 2013, 2016 and 2017. From 20 to 30 plants per accession were obtained. In 2013 four plants per accession and in 2016 and 2017 ten plants per accession were maintained. Wild species S. michoacanum (W type), cv. Early Rose (T type), breeding line PW 363 (D type), cultivated species S. phureja (P type) and cv. Maris Piper (A type) were used as a standard for multiplex PCR analysis [10]. As a standard for α, β, γ types cv. Nevsky, Early Rose and Stobrawa were used.
DNA extraction, PCR and restriction digestion
From 1 to 14 individual plants per accession were used for DNA extraction. Total DNA was extracted from 200 mg of fresh, young leaves of greenhouse-grown plant using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The DNA quality was checked with a NanoDrop Lite Spectrophotometer (Thermo Scientific) and assessed on 1% agarose gels. Genomic DNA was used in multiplex polymerase chain reaction (PCR) amplification with cpDNA- and mtDNA-specific primers and digested with BamHI restriction enzyme. Four chloroplast-specific markers (T, S, SAC, A) and one mitochondrial DNA marker (D) were used. MtDNA types α, ß and γ were distinguished using the ALM_4 and ALM_5 primers. To confirm the identification of M-type, an additional single PCR of marker A along with digestion was performed. Product amplification, restriction digestion and agarose gel electrophoresis were done according to Hosaka and Sanetomo [10] with modifications described in Smyda-Dajmund et al. [22]. Amplification reactions (20 µl reaction mixture) consisted of 2 µl 10 × buffer including 20 mM MgCl2 (Fermentas Life Sciences, Thermo Fischer Scientific, Waltham, Massachusetts, USA), 0.5 mM of each dNTP, 2 µM primer T, S and SAC and 3 µM primer D and A, 0.05 U/µl DreamTaq polymerase (Fermentas Life Sciences, Thermo Fischer Scientific, Waltham, Massachusetts, USA) and 30 ng DNA template. Amplifications were performed in a T3000 thermocycler (Biometra GmbH, Göttingen, Germany). The PCR parameters for multiplex PCR were 95 °C for 10 min followed by 35 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s and one final extension at 72 °C for 5 min. Digestion of the amplicons with restriction endonuclease BamHI (Fermentas Life Sciences, Thermo Fischer Scientific, Waltham, Massachusetts, USA) was performed according to producer’s protocol at 37 °C for 3 h. The PCR amplification of ALM_4 and ALM_5 [13] marker were prepared in volume 20 µl using 2 µl 10 × buffer including 20 mM MgCl2 Fermentas Life Sciences, Thermo Fischer Scientific, Waltham, Massachusetts, USA), 3 µM of each ALM_4 and ALM_5 primers, 0.05 U/µl DreamTaq polymerase (Fermentas Life Sciences, Thermo Fischer Scientific, Waltham, Massachusetts, USA) and 30 ng DNA template. The reaction conditions were incubation at 95 °C for 10 min followed by 35 cycles at 94 °C for 30 s, 57 °C for 60 s, 72 °C for 90 s and one final extension at 72 °C for 5 min. The products of multiplex PCR were separated in 1.5% high resolution agarose gels (EURx, Gdańsk, Poland). Amplicons of ALM_4 and ALM_5 marker were separated in 1.5% standard agarose gels. PCR products were visualized by ethidium bromide staining and were assessed under UV light after electrophoresis in 1 × TBE buffer (Tris–Borate-EDTA). A 100-bp DNA ladder (Invitrogen, Thermo Fischer Scientific, Waltham, Massachusetts, USA) was used to determine marker sizes.
Sequencing
For PCR product sequence analysis of T marker, four genotypes of S. famatinae (RUS001:4304; POL003:333139) were chosen. The PCR products were cut out from the agarose gel under UV light and purified with a GenElute Gel Extraction Kit (Sigma-Aldrich, St. Louis MO, USA) according to the manufacture’s protocol. DNA sequencing was done by Genomed S. A, Warsaw, Poland.
Results
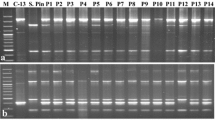
Cytoplasm types were determined for 43 accessions of 28 wild potato species, in total 401 genotypes (Table 1). In the accession of S. kurtzianum (RUS001:2301; POL003:333130) only one plant was obtained and preserved. From 1 to 14 genotypes per accession were analyzed. Twenty-two accessions had W cytoplasmic type, 17 had D – type and 4 had M-type. No segregation of cytoplasm types was observed within individuals of the same accession. From all 401 genotypes characterized, 201 (50.1%), 156 (38.9%) and 44 (11%) had cytoplasm types W, D, M, respectively (Table 1). No accessions with the T, P or A cytoplasm types were found. Within cytoplasm W, two subtypes W/α and W/β were identified. There was no accession identified with cytoplasm type W/γ. Different than standard marker pattern was observed within 14 genotypes of S. famatinae (RUS001:4304; POL003:333139). In order to determine which marker amplified in multiplex PCR in different way, single PCR for T and D markers were performed. An unusual profile was obtained with the T marker (Fig. 1). The obtained product was sequenced and a sequence of 502 bp was deposited in the National Center for Biotechnology Information (NCBI) GenBank database under the MN723848 accession number. Then the sequence was used to search for sequences of greatest similarity (i.e., the lowest e value and the greatest identity and maximum coverage) within the GenBank database. The closest matching sequences were found among the complete plastid genome sequences of wild and cultivated diploid potatoes belonged to Solanum, section Petota [27]. The query sequence was identical to the S. vernei PI320332 (sequence ID: NC_041633.1) plastid genome region spanning a range from 52,416 to 52,917 bps. Both sequences contained 65 bp insertion that differentiated them from standard products obtained with T marker, characteristic for W cytoplasm type.
Different amplification of T marker in multiplex PCR after BamHI digestion within S. famatinae (RUS001:4304; POL003:333139) individuals (samples 4–7). Samples 1–3 S. dolichostigma (RUS001:7613; POL003:333115); 8–11 S. fendleri (RUS001:5751; POL003:333112); 12–15 S. fendleri (RUS001:5747; POL003:333143); 16–18 S. fendleri (RUS001:5671; POL003:333110). Standards: wild species S. michoacanum (W), cv. Early Rose (T), breeding line PW 363 (D), cultivated species S. phureja (P) and cv. Maris Piper (A). T marker is marked with an arrow
Analysis based on multiplex PCR (T, S, SAC, A and D markers) indicated lack of segregation of cytoplasm types among individuals belonging to the same accession (Table 1). We also observed uniform cytoplasm type for the individuals of the same species with different accessions. As an example, five accessions of S. papita and four accessions of S. polytrichon were all with D cytoplasm type (Table 1). ALM_4/ALM_5 mtDNA primers generated five (type 0, 1, 2, 3, 4) different banding patterns among standards [10]. Band patterns of types 1, 2 and 3 were the only ones present among the analyzed accessions. Types 1, 2, 3 were observed when ALM_4/ALM_5 marker was used with the potato genotypes of W cytoplasm and 2, 3 among individuals with M cytoplasm type. All of the accessions of the cytoplasm D type had type 2 band pattern of the ALM_4/ALM_5 marker. We observed segregation of ALM_4/ALM_5 marker among individuals of S. leptophyes (RUS001:5764; POL003:333113). Out of 10 analyzed plants, five had profile 1 and five were with type 3 (Table 1).
Discussion
The study provides first data on cytoplasm diversity in a part of Vavilov potato collection multiplied in Poland. We have determined the types of cpDNA and mtDNA of 401 genotypes preserved in IHAR-PIB, based on a molecular marker system elaborated by Hosaka and Sanetomo [10]. We identified three cytoplasmic types: W (50.1%), D (38.9%) and M (11%) in the analyzed forms. Within type W subtypes W/α and W/β were identified. Fourteen genotypes of S. famatinae (RUS001:4304; POL003:333139) had W cytoplasm type, but with different amplicon of T marker. There was no sterilizing subtype W/γ. Type W and D dominated among tested genotypes and T-cytoplasm was not identified. Male-fertile cytoplasm types frequent in the material should be preserved and introduced into breeding lines in order to avoid fertility problems and widen the range of possible crosses in future generations. The dominance of a given type of cytoplasm depends on the types of materials stored in the genebank. Currently, the T type cytoplasm dominates among the potato cultivars and genebank resources. Japanese collection consists of 84 Japanese varieties and 378 breeding lines, 26 landraces and 260 foreign varieties and breeding lines [10]. The authors noticed dominance of cytoplasm T (73.9%) and D (17.4%). The frequency of cytoplasm W was slightly different than in other collections and was 2.4%. Domination of sterilizing cytoplasm types has also been noticed in other potato collections. From 978 genotypes of CIP collection 440 (45%), 368 (37.6%) and 110 (11.2%) had cytoplasm T, D and W, respectively [21]. Other analyzed genotypes had cytoplasm types: A, M and P with frequency 5.4%, 0.5% and 0.2%, respectively. The authors did not use ALM_4/ALM_5 marker and determination of sterilizing cytoplasm type W/γ was impossible, but they predicted its high frequency in CIP materials because of the intensive usage of S. stoloniferum as seed parent in breeding programs. 1,217 European potato cultivars and breeding clones maintained in German potato collection had T (59.4%), D (27.4%), W (12.2%) cytoplasm types. Remaining forms had A (0.7%) and M (0.3%) cytoplasm [17]. Cytoplasm types of 185 potato cultivars bred in Russia and Former Soviet Union countries were also described [28]. The authors have identified cytoplasm types: T, D and W/γ with frequency 40%, 50.8% and 8.7%, respectively. T cytoplasmic type is the most prevalent within S. tuberosum ssp. tuberosum, but it was also noticed in some forms of S. tuberosum ssp. andigena and diploid species of S. stenotomum [29,30,31]. It is not specific for wild potato species, but some accession of S. tarijense, S. berthaultii and S. neorossii were classified as T type [31]. Domination of T cytoplasm in European potatoes is caused by its origin from common ancestors: ‘Rough Purple Chili’, ‘Garnet Chili’ and ‘Early Rose’ and others S. tuberosum ssp. tuberosum clones [17, 32,33,34]. The second cytoplasm frequent in potato collections and also sterilizing is D-type. Its presence is associated with the introduction of resistance to P. infestans from S. demissum with cytoplasmic D-type in breeding programs. Cytoplasmic subtype W/γ was introduced into potato genepool from seed parents of S. stoloniferum carrying PVY resistance Rysto gene. This cytoplasm type was also found in S. chacoense, S. pampasense, S. pinnatisectum and S. vernei [10]. The choice of seed parent is crucial to reduce invasive sterilizing cytoplasm types.
T marker is an ndhlC/trn intergenic region in potato plastid genome [20]. The size of amplified fragment depends on potato species: approximately 440 bp fragment is specific for wild species or S. tuberosum ssp. andigena, a fragment of approximately 200 bp size is observed among S. tuberosum ssp. tuberosum, indicating a 241 bp deletion [31]. Hosaka [31] identified a longer product of T marker (of approximately 500 bp in size) among three accessions of S. vernei, with an insertion of 65 bp, a duplicated sequence of the position from 219 to 283 bp. We identified a PCR product of 502 bp in S. famatinae (RUS001:4304; POL003:333139), which was identical to the S. vernei PI320332 (sequence ID: NC_041633.1) plastid genome region spanning a range from 52,416 to 52,917 bps [27]. Both sequences contained 65 bp insertion that differentiated them from standard products obtained with T marker, characteristic for W cytoplasm type (437 bp in size). This was confirmed by comparing the obtained sequence with the plastid sequence of selected wild species deposited at NCBI by Huang et al. [27]. We also noticed that six S. vernei and two S. spegazzinii (synonym S. famatinae) accessions do not have an insertion in the T marker [27]. Both S. famatinae (synonym S. spegazzinii) and S. vernei come from Argentina, province Catamarca and Tucumán [4]. They are diploid wild species with EBN = 2. They belong to the same Tuberosa series, group Bolivia, Argentina and Chile according to Hawkes [4]. It is possible that this change occurred independently in both species, or they are the same species collected by independent collectors, misidentified and maintained in different genebanks. The argument for that hypothesis is their common geographical origin, but further analyses are necessary.
No segregation of markers T, S, SAC, A and D was observed both within the accessions and within species with multiple accessions. Hosaka and Sanetomo [10, 35] observed segregation of cytoplasm type within the same species. Lack of segregation of cytoplasm type in our material may result from the structure of VIR collection. Within S. leptophyes (RUS001:5764; POL003:333113), two types 1 and 3 of mitochondrial ALM_4/ALM_5 marker were observed. Segregation of this marker indicated that potato seed accessions are heterogeneous due to both nuclear and cytoplasm DNA polymorphisms. Hosaka and Sanetomo [10, 35] noticed segregation of cytoplasm types and marker ALM_4/ALM_5 within the species, but not within the same accession. The PCR reaction was carried out on DNA isolated twice from the same plants, but accidental mix of the seed material cannot be excluded.
Male-fertile cytoplasm types frequent in the material, if preserved and introduced into breeding lines, give a chance of avoiding fertility problems and widening the range of possible crosses in future generations of breeding materials.
Data availability
Plant material is maintained at Plant Breeding and Acclimatization Institute –National Research Institute in Poland.
Code availability
None.
References
Hosaka K (2003) T-type chloroplast DNA in Solanum tuberosum L. ssp. tuberosum was conferred from some populations of S. tarijense Hawkes. Am J Potato Res 80:21–32. https://doi.org/10.1007/BF02854553
Hawkes JG (1994) Origins of cultivated potatoes and species relationships. In: Bradshaw JE, Mackay GR (eds) Potato genetics. CAB International, Wallingford, pp 3–42
Machida-Hirano R (2015) Diversity of potato genetic resources. Breed Sci 65:26–40. https://doi.org/10.1270/jsbbs.65.26
Hawkes JG (1990) The potato, evolution, biodiversity and genetic resources. Belhaven Press, London
Spooner DM, Hijmans RJ (2001) Potato systematic and germplasm collecting, 1989–2000. Am J Potato Res 78:237–268. https://doi.org/10.1007/BF02875691
Spooner DM, Salas A (2006) Structure, biosystematics, and genetic resources. In: Gopal J, Khurana SMP (eds) Handbook of potato production, improvement, and post-harvest management. Haworth’s Press, Binghampton, pp 1–39
Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T (2014) Systematics diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev 80:283–383. https://doi.org/10.1007/s12229-014-9146-y
Birky CW Jr (1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA 92:11331–11338. https://doi.org/10.1073/pnas.92.25.11331
Lössl A, Adler N, Horn R, Frei U, Wenzel G (1999) Chondriome-type characterization of potato: mt α, ß, γ, δ, ε and novel plastid-mitochondrial configurations in somatic hybrids. Theor Appl Genet 98:1–10. https://doi.org/10.1007/s001220051202
Hosaka K, Sanetomo R (2012) Development of rapid identification method for potato cytoplasm and its use for evaluating Japanese collections. Theor Appl Genet 125:1237–1251. https://doi.org/10.1007/s00122-012-1909-4
Anisimova IN, Gavrilenko TA (2017) Cytoplasmic male sterility and prospects for its utilization in potato breeding, genetic studies and hybrid seed production. Russ J Genet Appl Res 7:721–735. https://doi.org/10.1134/S2079059717070024
Grun P (1979) Evolution of the cultivated potato: a cytoplasmic analysis. In: Hawkes JG, Lester RN, Skelding AD (eds) The biology and taxonomy of the Solanaceae. Academic Press, London, pp 655–665
Lössl A, Götz M, Braun A, Wenzel G (2000) Molecular markers for cytoplasm in potato: male sterility and contribution of different plastid-mitochondrial configurations to starch production. Euphytica 116:221–230. https://doi.org/10.1023/A:1004039320227
Hilali A, Fl L, Veilleux RE (1987) Reciprocal differences between hybrids of Solanum tuberosum Groups Tuberosum (haploid) and Phureja. Euphytica 36:631–639. https://doi.org/10.1007/BF00041513
Hoopes RW, Plaisted RL, Cubillos AG (1980) Yield and fertility of reciprocal-cross Tuberosum-Andigena hybrids. Am Potato J 57:275–284. https://doi.org/10.1007/BF02855305
Maris M (1989) Analysis of an incomplete diallel cross among three ssp. tuberosum varieties and seven long-day adapted ssp. andigena clones of the potato (Solanum tuberosum L.). Euphytica 41:163–182. https://doi.org/10.1007/BF00022425
Sanetomo R, Gebhardt C (2015) Cytoplasmic genome types of European potatoes and their effects on complex agronomic traits. BMC Plant Biol 15:162. https://doi.org/10.1186/s12870-015-0545-y
Sanford JC, Hanneman RE Jr (1979) Reciprocal differences in the photoperiod reaction of hybrid populations in Solanum tuberosum. Am Potato J 56:531–540. https://doi.org/10.1007/BF02855968
Sanford JC, Hanneman RE Jr (1982) Large yield differences between reciprocal families of Solanum tuberosum. Euphytica 31:1–12. https://doi.org/10.1007/BF00028300
Chimote VP, Chakrabarti SK, Pattanayak D, Padney SK, Naik PS (2008) Molecular analysis of cytoplasm type in Indian potato varieties. Euphytica 162:69–80. https://doi.org/10.1007/s10681-007-9563-7
Mihovilovich E, Sanetomo R, Hosaka K, Ordoñez B, Aponte M, Bonierbale M (2015) Cytoplasmic diversity in potato breeding: case study from the International Potato Center. Mol Breeding 35:137. https://doi.org/10.1007/s11032-015-0326-1
Smyda-Dajmund P, Śliwka J, Wasilewicz-Flis I, Jakuczun H, Zimnoch-Guzowska E (2016) Genetic composition of interspecific potato somatic hybrids and autofused 4x plants evaluated by DArT and cytoplasmic DNA markers. Plant Cell Rep 35:1345–1358. https://doi.org/10.1007/s00299-016-1966-2
Tiwari JK, Chandel P, Singh BP, Bhardwaj V (2014) Analysis of plastome and chondriome genome types in potato somatic hybrids from Solanum tuberosum x Solanum etuberosum. Genome 57:29–35. https://doi.org/10.1139/gen-2013-0122
Tiwari JK, Devi S, Chandel P, Ali N, Bhardwaj V, Singh BP (2016) Organelle genome analysis in somatic hybrids between Solanum tuberosum and S. pinnatisectum revealed diverse cytoplasm type in potato. Agric Res 5:22–28. https://doi.org/10.1007/s40003-015-0197-z
Zoteyeva N, Chrzanowska M, Flis B, Zimnoch-Guzowska E (2012) Resistance to pathogens of the potato accessions from the collection of N.I. Vavilov Institute of Plant Industry (VIR). Am J Potato Res 89:277–293. https://doi.org/10.1007/s12230-012-9252-5
Raman KV, Zimnoch-Guzowska E, Zoteyeva N (2000) The Vavilov Research Institute’s potato collection survived the siege of Leningrad—will it survive 21st-century economic restructuring? Diversity. News J Int Genet Resour Commun 16(3):12–15
Huang B, Ruess H, Liang Q, Colleoni C, Spooner DM (2019) Analyses of 202 plastid genomes elucidate the phylogeny of Solanum section Petota. Sci Rep 9:4454. https://doi.org/10.1038/s41598-019-40790-5
Gavrilenko TA, Klimenko NS, Alpatieva NV, Kostina LI, Lebedeva VA, Evdokimova ZZ, Apalikova OV, Novikova LY, Antonova OY (2019) Cytoplasmic genetic diversity of potato varieties bred in Russia and FSU countries. Vavilovskii Zhurnal Genetiki i Selektsii= Vavilov J Genet Breed 23(6):753–764. https://doi.org/10.18699/VJ19.534
Hosaka K, Hanneman RE Jr (1988) The origin of the cultivated tetraploid potato based on chloroplast DNA. Theor Appl Genet 76:172–176. https://doi.org/10.1007/BF00257842
Hosaka K (1995) Successive domestication and evolution of the Andean potatoes as revealed by chloroplast DNA restriction endonuclease analysis. Theor Appl Genet 90:356–363. https://doi.org/10.1007/BF00221977
Hosaka K (2002) Distribution of the 241 bp deletion of chloroplast DNA in wild potato species. Am J Potato Res 79:119–123. https://doi.org/10.1007/BF02881520
Plaisted RL, Hoopes RW (1989) The past record and future prospects for the use of exotic potato germplasm. Am Potato J 66:603–627. https://doi.org/10.1007/BF02853982
Provan J, Powell W, Dewar H, Bryan G, Machray GC, Waugh R (1999) An extreme cytoplasmic bottleneck in the modern European cultivated potato (Solanum tuberosum) is not reflected in decreased levels of nuclear diversity. Proc R Soc Lond B 266:633–639. https://doi.org/10.1098/rspb.1999.0683
Mendoza HA, Haynes FL (1974) Genetic relationships among potato cultivars grown in the United States. HortScience 9:328–330
Hosaka K, Sanetomo R (2009) Comparative differentiation in mitochondrial and chloroplast DNA among cultivated potatoes and closely related wild species. Genes Genet Syst 84:371–378. https://doi.org/10.1266/ggs.84.371
Acknowledgements
The authors thank Dr. Emil Stefańczyk (Plant Breeding and Acclimatization Institute – National Research Institute, Poland) for help with bioinformatics. The research was financed within G2P-SOL project (Title: Linking genetic resources, genomes and phenotypes of Solanaceous crops) that has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 677379 as well as statutory donation 1-3-00-3-05 and 3-1-02-0-01 from the Polish Ministry of Science and Higher Education and grant: UMO-2012/07/B/NZ9/01901financed by the National Science Centre in Poland.
Funding
The research was financed within G2P-SOL project (Title: Linking genetic resources, genomes and phenotypes of Solanaceous crops) that has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 677379 as well as statutory donation 1-3-00-3-05 and 3-1-02-0-01 from the Polish Ministry of Science and Higher Education and grant: UMO-2012/07/B/NZ9/01901financed by the National Science Centre in Poland.
Author information
Authors and Affiliations
Contributions
PSD: participated in the design of the study, molecular analysis of organellar DNA and drafted the manuscript; JŚ: participated in the design of the study and its coordination, took part in writing of the manuscript; MJ: maintenance of plant material, took part in writing; EZG: participated in the design of the study and its coordination, took part in writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Informed consent
Not applicable.
Research involving Human Participants and/or Animals
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Smyda-Dajmund, P., Śliwka, J., Janiszewska, M. et al. Cytoplasmic diversity of potato relatives preserved at Plant Breeding and Acclimatization Institute in Poland. Mol Biol Rep 47, 3929–3935 (2020). https://doi.org/10.1007/s11033-020-05486-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05486-4