Abstract
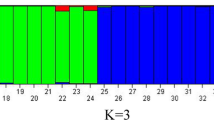
Evaluation of the genetic diversity in conventional and modern rapeseed cultivars is essential for conservation, management and utilization of these genetic resources for high yielding hybrid production. The objective of this research was to evaluate a collection of 86 oilseed rape cultivars with 188 simple sequence repeat (SSR) markers to assess the genetic variability, heterotic group identity and relationships within and between the groups identified among the genotypes. A total of 631 alleles at 188 SSR markers were detected including 53 and 84 unique and private alleles respectively, which indicated great richness and uniqueness of genetic variation in these selected cultivars. The mean number of alleles per locus was 3.3 and the average polymorphic information content was 0.35 for all microsatellite loci. Unweighted Pair Group Method with Arithmetic Mean clustering and principal component analysis consistently divided all the cultivars into four distinct groups (I, II, III and IV) which largely coincided with their geographical distributions. The Chinese origin cultivars are predominantly assembled in Group II and showed wide genetic base because of its high allelic abundance at SSR loci while most of the exotic cultivars grouped into Group I and were highly distinct owing to the abundant private and unique alleles. The highest genetic distance was found between Group I and IV, which mainly comprised of exotic and newly synthesized yellow seeded (1728-1 and G1087) breeding lines, respectively. Our study provides important insights into further utilization of exotic Brassica napus accessions in Chinese rapeseed breeding and vice versa.


Similar content being viewed by others
References
Becker HC, Loptien H, Robbelen G (1999) Breeding: an overview. In: Gomez-Campo C (ed) Biology of Brassica coenospecies. Elsevier, Amsterdam, pp 413–460
Salunkhe DK, Chavan JK, Adsule RN, Kadam SS (1992) World oilseeds, chemistry technology and utilization. Van Nostrand Reinbold, New York, p 59
Liu HL (1985) Rapeseed genetics and breeding. Shanghai Science and Technology Press, Shanghai, pp 38–42
Zhou WJ (2001) Oilseed rape. In: Zhang GP, Zhou WJ (eds) Crop cultivation. Zhejiang University Press, Hangzhou, pp 153–178
Fu T (2000) Breeding and utilization of rapeseed hybrid. Hubei Science Technology, Hubei, pp 167–169
Rakow G, Woods DL (1987) Outcrossing in rape and mustard under Saskatchewan prairie conditions. Can J Plant Sci 67:147–151
Snowdon RJ, Lühs W, Friedt W (2006) Oilseed rape. In: Kole C (ed) Genome mapping and molecular breeding, vol 2: oilseeds. Springer Verlag, Heidelberg, pp 55–114
Prakash S, Hinata K (1980) Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot 55:1–57
Downey RK (1964) A selection of Brassica campestris L. containing no erucic acid in its seed oil. Can J Plant Sci 44:499–504
Krzymanski J (1970) Inheritance of thioglucoside content by rapeseed (Brassica napus). Journees Internationales sur le Colza. C.E.T.I.O.M, 212–218
Seyis F, Snowdon R, Luhs W, Friedt W (2003) Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breed 122:473–478
Messmer MM, Melchinger AE, Herrmann RG, Boppenmaier J (1993) Relationships among early European maize inbreeds: II. Comparison of pedigree and RFLP data. Crop Sci 33:944–950
Lefort-Buson M, Guillot-Lemoine B, Dattée Y (1987) Heterosis and genetic distance in rapeseed (Brassica napus L.): crosses between European and Asiatic selfed lines. Genome 29:413–418
Gehringer A, Spiller T, Basunanda P, Snowdon R, Friedt W (2007) New oilseed rape (Brassica napus) hybrids with high levels of heterosis for seed yield under nutrient-poor conditions. Breed Sci 57:315–320
Diers BW, McVetty PBE, Osborn TC (1996) Relationship between heterosis and genetic distance based on restriction fragment length polymorphism markers in oilseed rape (Brassica napus L.). Crop Sci 36:79–83
Riaz A, Li G, Quresh Z, Swati MS, Quiros CF (2001) Genetic diversity of oilseed Brassica napus inbred lines based on sequence related amplified polymorphism and its relation to hybrid performance. Plant Breed 120:411–415
Ali M, Copeland LO, Elias SG, Kelley JD (1995) Relationship between genetic distance and heterosis for yield and morphological traits in winter canola (Brassica napus L.). Theor Appl Genet 91:118–121
Yu CY, Hu SW, Zhao HX, Guo AG (2005) Genetic distances revealed by morphological characters, isozymes, protein and RAPD markers and their relationships with Hybrid performance in oilseed rape (Brassica napus L.). Theor Appl Genet 110:511–518
Prasad M, Varshnez RK, Roy JK, Balyan HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Diers BW, Osborn TC (1994) Genetic diversity of oilseed Brassica napus germplasm based on restriction fragment length polymorphisms. Theor Appl Genet 88:662–668
Lombard V, Baril CP, Dubreuil P, Blouet F, Zhang D (2000) Genetic relationships and fingerprinting of rapeseed cultivars by AFLP: consequences for varietal registration. Crop Sci 40:1417–1425
Mailer RJ, Wratten N, Vonarx M (1997) Genetic diversity amongst Australian canola cultivars determined by randomly amplified polymorphic DNA. Aust J Exp Agric 37:793–800
Tommasini L, Batley J, Arnold GM, Cooke RJ, Donini P, Lee D, Law JR, Lowe C, Moule C, Trick M, Edwards KJ (2003) The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106:1091–1101
Hasan M, Seyis F, Badani A, Pons-Kühnemann J, Friedt W, Lühs W, Snowdon R (2006) Analysis of genetic diversity in the Brassica napus L. gene pool using SSR markers. Genet Resour Crop Evol 53:793–802
Powell W, Maachray GC, Proven J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Jones CJ, Edwards KJ, Castiglione S, Winfield MO, Sala F, Van de Weil AC, Bredemeijer G, Vosman B, Matthes M, Maly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linaacero R, Vazque A, Karp A (1997) Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3:381–390
Pejic I, Ajmore-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theor Appl Genet 97:1248–1255
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Blair MW, Diaz JM, Hidalgo R, Diaz LM, Duque MC (2007) Microsatellite characterization of Andean races of common bean (Phaseolus vulgaris L.). Theor Appl Genet 116:29–43
Charcosset AM, Lefort-Buson M, Gallais A (1991) Relationship between heterosis and heterozygosity at marker loci: a theoretical computation. Theor Appl Genet 81:571–575
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Deng W, Zhou L, Zhou YT, Wang YJ, Wang ML, Zhao Y (2010) Isolation and characterization of three duplicated PISTILLATA genes in Brassica napus. Mol Biol Rep. doi:10.1007/s11033-010-9981-9
Chen S, Nelson MN, Ghamkhar K, Fu T, Cowling WA (2008) Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51(1):1–10
Cheng X, Xu J, Xia S, Gu J, Yang Y, Fu J, Qian X, Zhang S, Wu J, Liu K (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor Appl Genet 118:1121–1131
Fan C, Cai G, Qin J, Li Q, Yang M, Wu J, Fu T, Liu K, Zhou Y (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor Appl Genet 121:1289–1301
Xu J, Qian X, Wang X, Li R, Cheng X, Yang Y, Fu J, Zhang S, King GJ, Wu J, Liu K (2010) Construction of an integrated genetic linkage map for the A genome of Brassica napus using SSR markers derived from sequenced BACs in B. rapa. BMC Genomics 11:594
Li H, Chen X, Yang Y, Xu J, Gu J, Fu J, Qian X, Zhang S, Wu J, Liu K (2010) Development and genetic mapping of microsatellite markers from whole genome shotgun sequences in Brassica oleracea. Mol Breed. doi:10.1007/s11032-010-9509-y
Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, Doucet I, Perret D, Villeger M, Vincourt P, Blanchard P (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111:1514–1523
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Oxford University Press, Oxford, pp 2128–2129
Rohlf FJ (2000) NTSYS-PC 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software, Setauket
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Junjian N, Colowit PM, Mackill D (2002) Evaluation of genetic diversity in rice subspecies by microsatellite markers. Crop Sci 42:601–607
Wang LX, Guan RX, Liu ZX, Chang RZ, Qiu LJ (2006) Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci 46:1032–1038
Gong X, Westcott S, Li C, Yan G, Lance R, Sun D (2009) Comparative analysis of genetic diversity between Qinghai–Tibetan wild and Chinese landrace barley. Genome 52:849–861
Zou J, Jiang C, Cao Z, Li R, Long Y, Chen S, Meng J (2010) Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome 53(11):908–916
Zhou WJ, Zhang GQ, Tuvesson S, Dayteg C, Gertsson B (2006) Genetic survey of Chinese and Swedish oilseed rape (Brassica napus L.) by simple sequence repeats (SSRs). Genet Resour Crop Evol 53:443–447
Butruille DV, Guries RP, Osborn TC (1999) Increasing yield of spring oilseed rape hybrids through introgression of winter germplasm. Crop Sci 39:1491–1496
Jones DF (1945) Heterosis resulting from degenerative changes. Genetics 30:527–542
Xiao J, Li J, Yuan L, McCouch SR, Tanksley SD (1996) Genetic diversity and its relationships to hybrid performance and heterosis in rice as revealed by PCR-based markers. Theor Appl Genet 92:637–643
Seyis F, Friedt W, Lühs W (2006) Yield of Brassica napus L. hybrids developed using resynthesised rapeseed material. Field Crops Res 96:176–180
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Acknowledgments
The research was supported by the National Natural Science Foundation of China (No. 31071452) and the Doctoral Fund of Ministry of Education of China (No. 20100146110019).
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Younas and Y. Xiao contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Younas, M., Xiao, Y., Cai, D. et al. Molecular characterization of oilseed rape accessions collected from multi continents for exploitation of potential heterotic group through SSR markers. Mol Biol Rep 39, 5105–5113 (2012). https://doi.org/10.1007/s11033-011-1306-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1306-0