Abstract
Wheat (Triticum aestivum L.) is a major staple crop in East Africa (EA) providing 9% and 10% of daily calories and protein intake, respectively. However, EA countries depend on import to meet 55% of their domestic wheat supplies due to increasing demands and low domestic yields. To determine the beneficial gene pool currently exploited for wheat improvement in EA, we examined the allelic diversity of 39 genes of breeding importance in a collection of 239 wheat cultivars and breeding lines from Kenya and Ethiopia using KASP markers. The assayed genes have been shown to control variations in plant height, thousand kernel weight (TKW), grain protein content, pre-harvest sprouting (PHS), disease resistance, and flowering time. We observed the beneficial alleles of some major genes including Gpc-B1, Yr5, Yr15, Sr26, and Fhb1 to be missing or present at low frequencies in this population. Furthermore, we validated the effects of the major Rht-1 alleles and TaCKX-D1 in controlling variation in plant height and thousand kernel weight, respectively, under EA conditions. Our results uncover hitherto unexploited allelic diversity that can be used to improve the genetic potential of EA wheat germplasm. This will inform strategies to rapidly mobilize these beneficial alleles for wheat improvement in EA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is one of the major cereals and staple crops in East Africa (EA). Due to the increasing rates of population growth and urbanization, as well as increases in household incomes in many EA countries, there has been a shift in dietary pattern to easy-to-cook meals including wheat-based diets (Negassa et al. 2013). This has led to a nearly two-fold increase in per-capita wheat consumption in EA over the last 60 years, at a time when per-capita consumption is declining in Europe. Wheat has therefore transitioned from a “colonial” crop confined to mission shambas (farms) to a food security crop accounting for 9% and 10% of daily calorie and protein intake in EA, respectively (FAOSTAT 2020). East Africa produces an average of 5 M tonnes of wheat per year from approx. 2 M ha of arable land (2014–2018 averages; FAOSTAT 2020). Up to 80% of wheat farmers in EA are small holder farmers, further underlying the importance of wheat production to household food security and livelihoods. Kenya and Ethiopia account for 92% of the wheat produced in EA; however, the average yield of wheat in both countries is low (~ 2 t/ha in 2017; FAOSTAT 2020). To meet the increasing demand for wheat, EA countries import 55% of their domestic wheat supplies. This heavy dependence on imports makes the region highly vulnerable to changes in global wheat supply market.
Advances in wheat genomics have enabled the identification of genes, and their allelic variations, that control many economically important traits in wheat. The development of molecular breeding approaches has also paved way for deploying beneficial alleles of these genes to accelerate wheat improvement in national and global wheat breeding programs. There is a long history of wheat breeding in Kenya and Ethiopia; however, the use of molecular breeding tools is very limited thereby hampering the rate of genetic gains achieved. As such, the national breeding programs in both countries have depended on introductions of wheat lines from international wheat breeding programs including CIMMYT and ICARDA. Understanding the composition and diversity of the gene pool in EA wheat germplasm is important for defining breeding strategies and prioritizing trait targets for wheat improvement.
Kompetitive Allele-Specific PCR (KASP) is regarded as a benchmark for SNP genotyping in marker-assisted breeding due to its low-cost, ease-of-use, specificity, and co-dominant nature (Semagn et al. 2014). Over the last decade, many functional KASP markers have been developed for genotyping allelic variations controlling economically important traits in global wheat germplasm. Functional genetic markers, unlike random genetic markers, are derived from allelic variations that have been shown to affect phenotypic variation by either association, linkage, or causation (Andersen and Lübberstedt 2003). There have been concerted efforts to assemble functional KASP markers in databases and publications making them easily accessible for marker-assisted breeding (CerealsDb: Wilkinson et al. 2012; MASWheat: https://maswheat.ucdavis.edu/). Recently, Rasheed et al. (2016) and Khalid et al. (2019) together describe the assembly of 127 functional KASP markers for studying allelic variation in 88 genes underpinning agronomic, adaptation, end-use quality, disease resistance, and drought tolerance traits.
The aims of this study were to (1) use readily-available KASP markers to evaluate the allelic diversity of genes of breeding importance, (2) examine the changes in the allele frequencies of these genes over the last century, and (3) evaluate the phenotypic effect of some of these genes on plant height and grain size variations under EA field conditions.
Materials and methods
Plant materials and phenotyping
A set of 239 bread wheat genotypes comprised of 150 lines from Ethiopia and 89 lines from Kenya was used. The Ethiopian wheat genotypes were obtained from the Ethiopian Institute of Agricultural Research (EIAR) and comprised 53 wheat cultivars released from 1970 to 2017 and 97 breeding lines currently under development. The Kenyan wheat genotypes comprised cultivars released from 1920 to 2012 and were obtained from the Kenya Agricultural and Livestock Research Organization (KALRO).
Three phenotyping experiments (Experiment 1–3) were conducted in Kenya and Ethiopia across 3 years. Experiments 1 and 2 were conducted independently for Kenyan and Ethiopian genotypes, respectively, to account for local environmental effects on the performance of the genotypes from each country. Experiment 1 was conducted at the EIAR Kulumsa Agricultural Research Centre (8° 01′ N, 39° 09′ E; 2200 m above sea level) during the 2017 main cropping season from July to November. Experiment 2 was conducted at KALRO Field Crop Research Station Njoro (0° 20′ S, 35° 56′ E; 2185 m above sea level) during the 2018 main cropping seasons from May to October. Experiment 3 comprised both Kenyan and Ethiopian genotypes and was conducted at the KALRO Field Crop Research Station in 2019. Experiments 1 and 3 in Kenya were set up using Randomized Complete Block design with two replications, while Experiment 2 in Ethiopia was set up using Alpha Lattice design with two replications. Each replication was sown in 1 m double-row plots.
Plant height measurements were obtained across all three experiments. Plant height was measured at physiological maturity from the plant base to the tip of the spike (excluding awns) of the primary tiller. For Experiments 1 and 2, each plot was hand-harvested at maturity, threshed, and a sample of 1000 kernels was taken and weighed to determine the thousand kernel weight (TKW).
The year of release and phenotypic data for each genotype are presented in Table S1 in Online Resource 1.
DNA extraction and KASP genotyping
Leaf samples were collected from a single plant for each line at the seedling stage. Genomic DNA was extracted following the CTAB method (Doyle and Doyle 1987) and ZYMO kits (ZR Plant/Seed DNA mini-Prep kit; Zymo Research, USA). The quality and quantity of DNA extracted were evaluated using spectrophotometric (NanoDrop™ 8000) and gel-based methods. All samples were diluted to a uniform concentration, and an equivalent of 50 ng of DNA per sample was used for genotyping.
KASP genotyping was done at the BecA-ILRI Hub (Kenya). A total of 59 KASP assays were used in this study. This comprises 55 of the 127 assays reported by Rasheed et al. (2016) and Khalid et al. (2019). We also obtained assay information for TaMKK3-A, Yr15 (Kin2), Yr5, and Sr26 directly from the publications describing the development of KASP markers for these genes (Shorinola et al. 2017; Klymiuk et al. 2019; Marchal et al. 2018; Qureshi et al. 2018). Each assay comprises of two allele-specific primers tailed with either FAM or HEX dye sequences and a common untailed primer. The gene information, primer sequences, and allele-dye assignment for each marker used are presented in Table S2 in Online Resource 1. The RefSeqv1 gene ID of each gene examined as well as the physical position of the primer sequences was obtained based on the best BLAST hit to the Chinese Spring Refseqv1 assembly hosted on Ensembl Plants (Table S2). The primers for each assay were mixed in the following proportion: 46 μL de-ionized water, 30 μL common primer (100 μM), and 12 μL of each tailed primer (100 μM). KASP genotyping was performed following the manufacturer’s (LGC Biosearch Technologies, UK) protocol. The assays were run in 384-well plate format. A reaction volume of 5.0 μL comprising 2.5 μL of DNA and 2.5 μL of a mix of 2x KASP reagent and 0.07 μL primer mix was used for amplification. PCR cycling was performed using the following protocol: hot start at 94 °C for 15 min, followed by ten touchdown cycles (94 °C for 20 s; touchdown at 61 °C initially and decreasing by − 1 °C per cycle for 60 s), followed by 30 cycles of annealing (94 °C for 60 s; 55 °C for 60 s). Additional cycles were performed to increase the intensity of the fluorescence signals where necessary. For each assay, we included no-template (water) controls as well as positive varietal controls that harbor alternate alleles of each gene (where available). Fluorescence signals from amplified samples were measured at < 40 °C using the Fluostar Omega Microplate Reader (BMG Labtech, UK). Genotype clustering was done using the KlusterCaller software (LGC Biosearch Technologies, UK).
Statistical analyses
Statistical analyses were done in R 3.6.0 (R Core Team 2019) using the LmerTest and car packages (Kuznetsova et al. 2017; Fox and Weisberg 2019). Only allelic variations with minimum allele frequency > 0.05 were included for statistical analyses. As plant height measurements were taken in at least two experiments for each genotype across three years, a linear mixed-effect function using Restricted Maximum Likelihood model was used to assess the effect of Rht-1 genes on plant height. For this, allelic variation of Rht-1 genes, year (2017–2019), and location (Kenya or Ethiopia) were taken as fixed effects, while genotype was taken as a random effect to account for the background genetic effects from each genotype. TKW measurement was only taken at one location (in Experiment 1 or 2) for each genotype. A linear model taking allelic variation for the grain size genes and location (Kenya or Ethiopia) as fixed effects was, therefore, used to assess the effect of grain size related genes on TKW.
Results
Allele frequencies of functional genes in EA wheat population
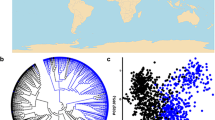
We genotyped 239 EA bread wheat cultivars and breeding lines using 59 KASP assays. Of these 59 assays, we obtained good genotype clusters (distinct from the no-template control) for 48 assays. These 48 assays target 39 genes/loci controlling plant height, drought tolerance, grain quality, grain size, flowering time, and disease resistance. Genotype information of all 48 working assays are presented in Table S3 in Online Resource 1. The remaining assays either produced clusters that were difficult to separate or had high proportions of samples with no amplification. Of the 48 working assays, 14 assays produced only one distinct genotype cluster separate from the no-template control (Fig. 1 a and b) suggesting that the germplasm are fixed for a single allele. We considered a gene allele to be favorable if it is causal, associated or linked to a desired agronomic, disease resistance, grain size, or end-use quality traits. In general, hard-grained, high-yielding, early-flowering, and semi-dwarf wheat varieties with good disease resistance attract higher premium in EA wheat market, as such gene alleles promoting these traits are considered favorable for wheat cultivation in EA (Fig. 2, see details in Table S2). Ten of the 14 single-cluster assays were fixed for unfavorable gene alleles, thus highlighting immediate opportunities for improving these traits in the EA wheat population. Some of the major genes fixed for unfavorable gene alleles in the EA wheat population include Yr5, Fhb1, and Sr26.
Allele frequencies of functional KASP markers in EA population. Allele frequencies of 48 markers for height, disease resistance, grain quality, grain size and flowering time genes in EA population. Polymorphism information for each gene is described on the left. The frequencies of favorable and unfavorable gene alleles are represented with green and light-brown colored bars, respectively. The allele nomenclature for each gene are indicated next to each bar
On the other hand, 34 assays produced two distinct genotype clusters suggesting segregation of favorable and unfavorable alleles (Fig. 1c). Of these segregating assays, 14 assays showed higher frequencies for the favorable alleles, while 17 showed higher frequencies for the unfavorable alleles (Fig. 2). Three assays showed near-balanced frequencies between the favorable and unfavorable alleles.
Allele frequency distribution for most of the genes examined showed a similar pattern in the Ethiopian and Kenyan germplasm in terms of pre-dominance of the favorable or unfavorable alleles (Fig. S1, Online Resource 2). This is most likely due to the common sources of founder materials and the frequent exchange of germplasm between the national wheat breeding programs of both countries. The only exceptions were for Rht-B1 and Pina-D1. The favorable allele of Rht-B1 was pre-dominant (86%) in Ethiopia compared to only 21% in Kenya. Similarly, the favorable allele of Pina-D1 was pre-dominant (85%) in Ethiopia compared to that (only 37%) in Kenya. The difference in allele frequencies for Rht-B1 is most likely due to the composition of the wheat collection from both countries in our study: 48% of the Kenyan collection were released before the introduction of Rht-1 genes during the introduction of the Green Revolution-derived cultivars in EA, while the Ethiopian collection comprised entirely of post-Green Revolution lines.
Changes in allele frequency distribution across breeding periods
The germplasm examined were introduced to or developed in EA across a 100-year period (1920 to 2017) spanning different breeding periods with access to different gene alleles and breeding technologies. As such, we further examined the changes in allele frequency distribution across these breeding periods for assays showing segregation. Four distinct wheat breeding periods were defined: pre-1970, before the introduction of the Green Revolution wheat cultivars to EA; 1970–2000, characterized by the introduction of Green Revolution derived germplasm into EA; 2001–2017, characterized by the use of molecular breeding tools; and breeding lines that are yet to be released. Due to their similar allele frequency distribution, the Kenyan and Ethiopian germplasm released in these breeding periods were considered together. We focused mainly on the distribution of allelic variations that are either causal or linked to target phenotypes as these are most likely to have contributed to trait variations and breeders selection compared to associated allelic variants that are yet to be validated. The distribution of associated allelic variations (mainly for grain size genes) are presented in Fig. S2 (Online Resource 2).
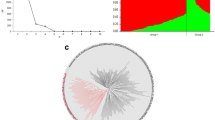
Plant height genes (Fig. 3): Four assays for the major reduced plant height gene alleles, Rht-B1 (Rht-B1a: tall; Rht-B1b: semi-dwarf), Rht-D1 (Rht-D1a: tall; Rht-D1b: semi-dwarf), and the 167-bp and 190-bp InDel polymorphisms in Rht-B1 were examined (Ellis et al. 2002; Wilhelm et al. 2013). Rht-B1 was the pre-dominant gene conferring reduced plant height in the EA wheat population, with 60% of EA wheat lines harboring the favorable semi-dwarf Rht-B1b allele compared to only 10% of EA lines containing the semi-dwarf Rht-D1b allele. Furthermore, the allele frequencies of Rht-B1b increased in lines released post-1970 with a drastic increase in lines released from 2001. While the allele frequency of the favorable semi-dwarf Rht-D1b initially increased in lines released post-Green Revolution, the frequencies subsequently decreased sharply in lines released since 2001. None of the genotypes examined harbor Rht-D1b and Rht-B1b alleles together. The 167-bp and 190-bp polymorphisms show no substantial change in allele frequency distribution across the breeding periods.
Grain quality genes (Fig. 4): Six grain quality genes showed progressive changes in allele frequency distributions across the breeding periods. These included the puroindolines (Pins) genes, Pina-D1 and Pinb-D1, which together control grain hardness (Morris 2002). Due to the higher premium for hard wheat grains compared to soft wheat grains, alleles that promote grain hardness (Pina-D1b and Pinb-D1b) were considered favorable relative to alleles for grain-softness (Pina-D1a and Pinb-D1a). Grain hardness is determined by the presence of either or both Pina-D1b and Pinb-D1b alleles. We observed a steady increase of the favorable Pina-D1b allele across the breeding periods from 15% in lines released before 1970 to 90% in current breeding lines. Conversely, the frequency of the favorable Pinb-D1b allele decreased across the breeding periods from 33% in lines released before 1970 to 6% in breeding lines. We also examined the distribution of the Pina-D1/Pinb-D1 haplotype across the breeding periods (Fig. S3, Online Resource 2). The soft-grain Pina-D1a/Pinb-D1a haplotype decreased steadily across the breeding periods as did one of the hard-grain haplotypes (Pina-D1a/Pinb-D1b). In contrast, the Pina-D1b/Pinb-D1a hard-grain haplotype increased across the breeding periods and is the pre-dominant haplotype in the breeding lines.
TaMFT-A1 (same as TaPhs-A1) and TaMKK3-A are two important genes controlling embryo-imposed seed dormancy in wheat (Vetch et al. 2019). Optimized seed dormancy level is advantageous for preventing pre-harvest sprouting (PHS) without compromising uniform seed germination. Different allelic variation exists for TaMFT-A1 including a promoter polymorphism (− 222 bp C/T; Nakamura et al. 2011), which is fixed in the EA population, and two segregating exonic and splice junction polymorphisms (A666T and G646A; Liu et al. 2013). Only one causal natural allelic variation has been reported for TaMKK3-A (Torada et al. 2016). We found higher frequencies of dormancy-promoting alleles of TaMFT-A1 compared to TaMKK3-A in the EA wheat population. The frequencies of the dormancy-promoting TaMFT-A1 and TaMKK-3A alleles however similarly reduced progressively over the breeding periods.
Other segregating grain quality genes examined included Gpc-B1 (grain protein content) and Glu-D1 (gluten strength). The high-protein Gpc-B1 introgression was missing in varieties released before 2000. It is, however, present at low frequencies in varieties released post-2000 (2%) and in breeding lines (4%). We also observed an increasing trend for the strong-gluten Glu-D1 (Dx5 + Dy10) alleles from 51% in pre-1970 lines to 91% in breeding lines.
Disease resistance genes (Fig. 5): Only six of the 11 disease-related allelic variations examined showed segregation. These included allelic variations for Yr15 for yellow rust resistance, Lr34 and Lr21 for leaf rust resistance, Tsn1 for tan spot sensitivity, and Cre8 for nematode resistance. Two allelic variations for Yr15 were examined including the causal Yr15_Kin2 (Klymiuk et al. 2019) and the linked Yr15_R8 (Ramirez-Gonzalez et al. 2015) polymorphisms. Although showing slightly different distributions, both Yr15_Kin2 and Yr15_R8 showed low frequencies of the favorable resistant +Yr15 allele across the breeding periods (0–23%). The distribution is however different for Lr34 (TTC InDel; Lagudah et al. 2009) and Tsn1 (Tsn1+/Null; Faris et al. 2010) which both showed pre-dominant and increasing frequencies of their resistant alleles across the breeding periods. The resistant +Lr21 gene allele was only present in one breeding line, and the resistant +Cre8 allele frequency was stably low (2–9%) across the breeding periods.
Flowering time genes: EA wheat germplasm mainly comprises photoperiod-insensitive spring wheat genotypes which are likely fixed for vernalization and photoperiod insensitivity gene alleles. We, therefore, focused on the earliness per se locus, Eps-D1, and its homoeologous locus Eps-B1, which also account for variation in flowering time irrespective of vernalization and photoperiod requirements (Zikhali et al. 2016; Alvarez et al. 2016). A large chromosomal deletion (including TaElf3-D1) and a SNP variation in TaElf3-D1 have been linked to the Eps-D1 effect (Zikhali et al. 2016). The chromosomal deletion is flanked by the TaBradi2g14790 marker. We observed a near-balanced allele frequency distribution for the chromosomal deletion and wild-type (non-deleted) alleles across the breeding periods (Fig. S4, Online Resource 2). This deletion variation was also confirmed by the fact that lines showing the chromosomal deletion showed no amplification for TaElf3-D1 (Fig. S5, Online Resource 2). The Elf3-B1 SNP variation linked to the Eps-B1 locus was also segregating in the EA population. We observed higher frequencies of the favorable early-flowering Elf3-B1 allele compared to the late-flowering alleles across the breeding periods with an increasing trend observed from cultivars released from 1970 to modern breeding lines (Fig. S4, Online Resource 2).
Field effect of allelic variations in plant height and grain size genes
We evaluated the phenotypic effects of the allelic variations in plant height and grain size genes in field trials conducted in Ethiopia and Kenya. From these trials, we confirmed the effect of the Green Revolution Rht-B1b and Rht-D1b semi-dwarfing alleles in reducing plant height. The semi-dwarf alleles of both genes are estimated to significantly (P < 0.0001) reduced height by 13.50 (± 1.55) cm and 14.93 (± 2.23) cm, respectively (Table 1). The Rht-B1 160 bp InDel variation did not show any significant effect on plant height in the EA population. The Rht-B1 190 bp InDel was almost fixed in the population (99.15%), as such its effect could not be statistically tested. We similarly examined the effect of seven segregating grain size genes including TaGS-D1, TaCKX-D1, TaCwi-4A, TaSus1-7B, TaGS5-A1, TEF-7A, and TaGS1a. Of these genes, only TaCKX-D1 showed a significant (P < 0.0001) effect on TKW with the deletion TaCKX-D1a allele estimated to increase TKW by 14.27 (± 1.99) g (Table 1). However, TaCKX-D1 only segregates in the Ethiopian population and is fixed for the favorable higher-TKW allele in the Kenyan population.
Discussions
We examined the allelic diversity of 39 genes affecting important agronomic, disease resistance, grain size, and end-use quality traits in EA wheat population. Most of these genes segregate in the EA population suggesting the presence of favorable and unfavorable alleles, although in different proportions. These highlight opportunities for wheat improvement in EA by using marker-assisted breeding to increase the frequencies of favorable gene alleles.
Opportunities to introduce major but missing genes in EA germplasm
Some of the major genes missing in EA wheat population include Sr26, Yr5, and Fhb1. Rusts (stem, yellow, and leaf) are major constraints to wheat production in EA. Kenya and Ethiopia are hotspots for highly virulent and rapidly evolving races of the stem rust pathogen (Puccinia graminis f. sp. tritici). This includes the Ug99 race (TTKSK) which was first discovered in Uganda in 1998 (Pretorius et al. 2000; Singh et al. 2011) and was virulent to most of the stem rust resistance genes deployed in global wheat germplasm at that time. About four additional variants of Ug99 each with increasing virulence to more stem rust resistance genes and five new non-Ug99 races have since been reported in Kenya and Ethiopia (Singh et al. 2015; Bhavani et al. 2019). Sr26, a gene introgressed from Agropyron elongatum (Liu et al. 2010), is one of very few resistance genes still effective against all known Ug99 variants. However, its use in global germplasm has been limited as a result of an associated yield penalty caused by linkage drag (Liu et al. 2010). This yield penalty possibly accounts for the lack of Sr26 in EA wheat population, although there is no evidence to suggest that Sr26 was ever introduced or tested under EA conditions. Lines with smaller introgression of Sr26 without the yield loss penalty (Dundas et al. 2007) have been developed, thus providing an opportunity for deploying and evaluating the effect of Sr26 on rust resistance and yield under EA conditions.
Similarly, Yr5 is one of very few genes conferring resistance against a wide range of virulent strains of the yellow rust pathogen (Puccinia striiformis f. sp. tritici; Marchal et al. 2018). Unlike Sr26, Yr5 is currently used in many breeding programs including in Europe and has no associated yield penalty. There should, therefore, be no disincentive against the use of Yr5 in EA germplasm. The infrequent occurrence of Fusarium Head Blight (FHB) in EA might account for the lack of Fhb1 in EA germplasm. Nonetheless, FHB still causes a significant yield loss and poses health risks due to the production of hazardous mycotoxins. There is, therefore, an opportunity to anticipatorily breed for resistance against the sporadic occurrence of FHB.
It is worth noting that we only examined disease resistance genes for which KASP markers have been developed. We cannot, therefore, exclude the presence of other functional resistance genes, particularly for stem and yellow rust, in the EA wheat population. The development and public deposition of more breeder friendly KASP marker assays for resistance genes will facilitate the examination and deployment of these genes in global germplasm. Also, the use of KASP markers to rapidly verify the absence of known resistance genes in otherwise resistant germplasm will highlight the presence of novel resistance genes in such germplasm and facilitate their identification.
Allele frequency changes across breeding periods and balancing selection for optimizing traits expression
We also examined changes in allele frequency distribution across different breeding periods for segregating genes alleles. This analysis highlights the progress made in increasing the favorable alleles of some genes (including Rht-B1, Glu-D1, Pina-D1, Tsn1, and Lr34) across the breeding periods. For genes like Rht-B1 and Lr34 whose effects are easily observed, the progressive increase in favorable allele frequencies might be due to direct phenotypic selection. The advent of breeder friendly molecular markers in wheat breeding might have also accelerated the selection of these alleles. For instance, the drastic increase in the semi-dwarf (favorable) allele of Rht-B1 in the lines released between 2001 and 2017 coincided with the development of “perfect” molecular markers for the Rht-1 genes by Ellis et al. (2002).
Our study also highlights the progressive loss of the favorable alleles of some genes: Rht-D1, Pinb-D1, and TaMKK-3A. While it is essential to work towards increasing the favorable alleles of these genes, it is important to bear in mind that the overall goal of an effective breeding program should be on traits improvement (i.e., improving favorable trait expression) rather than “allele” improvement per se (i.e., increasing favorable gene allele frequency). This is particularly important for multigenic traits where interactions (additive or epistatic) between the different genes controlling such traits are more important in determining the trait expression than the individual gene actions. Balancing the selection of interacting genes is therefore important for fine-tuning and optimizing the expression of such traits for different environments and end-uses. We found evidence for such balancing selection in EA germplasm. For instance, Rht-B1 and Rht-D1 additively control plant height, with plants having semi-dwarf alleles of both genes showing severely reduced plant height (dwarf). This dwarf phenotype is, however, not considered beneficial in Kenya and Ethiopia because it markedly reduces grain size and yield (Pinthus and Levy 1983; Inagai and Tahir 1991), and also reduces the amount of straws farmers can harvest for feeding livestock. Consequently, we observed that increase in the Rht-B1b allele is accompanied by a decrease in Rht-D1b allele. It is not exactly clear why the Rht-B1b allele was preferentially selected instead of Rht-D1b allele given that both alleles have similar effect on plant height and grain size. It is worth noting, however, that majority of the EA wheat varieties released post-1970 are introductions from international wheat breeding programs including CIMMYT and ICARDA (Macharia and Ngina 2017). In agreement with our result, Zhao et al. (2019) showed a higher percentage of the Rht-B1b allele in CIMMYT germplasm compared to the Rht-D1b allele. One possible reason for this low frequency of Rht-D1b allele is its association with reduced Type 2 FHB resistance compared to the Rht-B1b allele (Srinivasachary et al. 2008; Buerstmayr and Buerstmayr 2016).
We observed a similar pattern of balancing selection for TaMKK3-A and TaMFT-A1 which both control seed dormancy levels in an additive manner. Although the dormancy-promoting allele frequencies of both genes showed an overall decreasing trend, we observed that high frequency of the dormancy-promoting TaMFT1 allele is balanced by low frequency of the dormancy-promoting TaMKK3-A allele. Only 13% of lines examined contains dormancy-promoting alleles of both genes. This balanced selection ensures that an optimum level of dormancy is expressed that protects against PHS without impairing uniform seed germination.
Some of the genes/loci we examined, including Gpc-B1, Yr15, Pm21, Sr26, 1A/1R, 1B/1R, and Lr21, were introduced into bread wheat from progenitors and wild wheat species. Most of these “alien” introgressions are either absent (Lr21, 1A/1R, Sr26) or present at low frequencies (1B/1R, Gpc-B1, Yr15) in EA wheat population. Of the 239 lines examined, only five harbor Gpc-b1 and three of these (Kenya Tai, ETBW8725, ETBW8923) have Triticum dicoccom accessions in their pedigree — supporting the emmer origin of Gpc-B1 (Uauy et al. 2006). More than 97% of the lines containing the 1B/1R introgression were released post-1970. The 1B/1R introgression from rye has been an important source of disease resistance genes including Sr31 which was widely used for stem rust resistance in germplasm distributed by CIMMYT, including to EA, before the emergence of the Ug99 race (Johansson et al. 2020). The 1B/1R introgression is, however, linked with poor grain quality for bread making which perhaps accounts for its low frequency in EA population. A more extensive pedigree analysis of the EA collection along with genotyping of the major founder lines in these pedigrees will be required to confidently ascertain the exact origins of these introgressions in EA population.
Importance of validating population-specific effect of beneficial genes
Some of the markers investigated in this study, particularly for grain size genes, were identified by association analyses in wheat populations that are genetically distinct to the EA wheat population used in this study. It was, therefore, important to validate the effect of such genes in EA germplasm under local field conditions. Out of the seven grain size-related genes segregating in the EA wheat population, only TaCKX-D1 showed a significant effect on TKW. It is important to note that the polymorphism reported in these genes were only shown to be associated with, but were not validated as being causal of the natural variation in TKW (Zhang et al. 2012; Guo et al. 2013; Hou et al. 2014; Zheng et al. 2014; Jiang et al. 2015; Wang et al. 2016). It is, therefore, possible that the lack of phenotypic effect in our study might be due to difference in the haplotype structure between the EA wheat population and the original study population used to identify these markers. Such haplotype differences would not have been picked up through single-marker analysis for each gene as was done in our study. Grain size traits are also highly environmental dependent (Brinton and Uauy 2019), and it is possible that differences in environmental conditions might also have contributed to the lack of effects of some of the grain size genes examined. Similarly, Sukumaran et al. (2018) did not find a consistent significant effect for a reported functional allelic variation in an important grain size gene, TaGW2-A, on TKW when tested under 10 environments. On the other hand, causal polymorphisms in Rht-B1 and Rht-D1 which have been widely validated across global germplasm showed significant effects in our study. We therefore propose that haplotype-based analysis (involving the use of markers defining haplotype blocks around a gene of interest) should be used in validating the effect of associated genes under local growth conditions before using such genes for marker-assisted breeding.
Outlook
Knowledge of the allelic diversity currently used or missing in EA wheat population is important in informing strategies for wheat breeding in Kenya and Ethiopia. This is particularly important for combating diseases that are critically limiting wheat production in both countries. To this end, we are currently working towards introducing beneficial alleles of Yr5 (missing), Yr15 (low frequency), Sr26 (missing), and Sr22 in combination in a common locally adapted EA wheat line. We will also be introducing functional Fhb1 (missing) alleles to this background to future proof the developed lines against the sporadic occurrence of FHB.
Data availability
The genotype and phenotype data generated in this study are provided in the Online Resource 1 (supplementary tables).
References
Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J (2016) Genetic and physical mapping of the earliness per se locus Eps-Am1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomics 16:365–382. https://doi.org/10.1007/s10142-016-0490-3
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560. https://doi.org/10.1016/j.tplants.2003.09.010
Bhavani S, Hodson D, Huerta-Espino J et al (2019) Progress in breeding for resistance to Ug99 and other races of the stem rust fungus in CIMMYT wheat germplasm. Front Agr Sci Eng 6:210–224
Brinton J, Uauy C (2019) A reductionist approach to dissecting grain weight and yield in wheat. J Integr Plant Biol 61:337–358. https://doi.org/10.1111/jipb.12741
Buerstmayr M, Buerstmayr H (2016) The semidwarfing alleles Rht-D1b and Rht-B1b show marked differences in their associations with anther-retention in wheat heads and with Fusarium head blight susceptibility. Phytopathology 106:1544–1552. https://doi.org/10.1094/PHYTO-05-16-0200-R
Core Team R (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dundas IS, Anugrahwati DR, Verlin DC, Park RF, Bariana HS, Mago R, Islam AKMR (2007) New sources of rust resistance from alien species: meliorating linked defects and discovery. Aust J Agric Res 58:545–549. https://doi.org/10.1071/AR07056
Ellis H, Spielmeyer W, Gale R et al (2002) “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042. https://doi.org/10.1007/s00122-002-1048-4
FAOSTAT (2020) Statistical Database of the Food and Agriculture of the United Nations. http://www.fao.org/faostat/en/#home. Accessed 19 Apr 2020
Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS, Oliver RP, Simons KJ, Friesen TL (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci U S A 107:13544–13549. https://doi.org/10.1073/pnas.1004090107
Fox J, Weisberg S (2019) An R companion to applied regression. Third, Sage, Thousand Oaks
Guo Y, Sun J, Zhang G, Wang Y, Kong F, Zhao Y, Li S (2013) Haplotype, molecular marker and phenotype effects associated with mineral nutrient and grain size traits of TaGS1a in wheat. Field Crop Res 154:119–125. https://doi.org/10.1016/j.fcr.2013.07.012
Hou J, Jiang Q, Hao C, Wang Y, Zhang H, Zhang X (2014) Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol 164:1918–1929. https://doi.org/10.1104/pp.113.232454
Inagai M, Tahir M (1991) Effects of semi-dwarfing genes Rht1 and Rht2 on yield in doubled haploid lines of wheat. Jpn J Breed 41:163–167. https://doi.org/10.1270/jsbbs1951.41.163
Jiang Y, Jiang Q, Hao C, Hou J, Wang L, Zhang H, Zhang S, Chen X, Zhang X (2015) A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor Appl Genet 128:131–143. https://doi.org/10.1007/s00122-014-2417-5
Johansson E, Henriksson T, Prieto-Linde ML et al (2020) Diverse wheat-alien introgression lines as a basis for durable resistance and quality characteristics in bread wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2020.01067
Khalid M, Afzal F, Gul A et al (2019) Molecular characterization of 87 functional genes in wheat diversity panel and their association with phenotypes under well-watered and water-limited conditions. Front Plant Sci 10:717. https://doi.org/10.3389/fpls.2019.00717
Klymiuk V, Fatiukha A, Fahima T (2019) Wheat tandem kinases provide insights on disease-resistance gene flow and host–parasite co-evolution. Plant J 98:667–679. https://doi.org/10.1111/tpj.14264
Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmerTest package: tests in linear mixed effects models. J Stat Softw 82:1–26. https://doi.org/10.18637/jss.v082.i13
Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guedira G, Selter LL, Keller B (2009) Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet 119:889–898. https://doi.org/10.1007/s00122-009-1097-z
Liu S, Yu L-X, Singh RP, Jin Y, Sorrells ME, Anderson JA (2010) Diagnostic and co-dominant PCR markers for wheat stem rust resistance genes Sr25 and Sr26. Theor Appl Genet 120:691–697. https://doi.org/10.1007/s00122-009-1186-z
Liu S, Sehgal SK, Li J, Lin M, Trick HN, Yu J, Gill BS, Bai G (2013) Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195:263–273. https://doi.org/10.1534/genetics.113.152330
Macharia G, Ngina B (2017) Wheat in Kenya: past and twenty-first century breeding. Wheat Improv Manag Util. https://doi.org/10.5772/67271
Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM, Boyd L, McIntosh R, Wulff BBH, Berry S, Lagudah E, Uauy C (2018) BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants 4:662–668. https://doi.org/10.1038/s41477-018-0236-4
Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48:633–647. https://doi.org/10.1023/A:1014837431178
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23:3215–3229. https://doi.org/10.1105/tpc.111.088492
Negassa A, Shiferaw B, Koo J, et al (2013) The potential for wheat production in Africa: analysis of biophysical suitability and economic profitability. Mexico
Pinthus MJ, Levy AA (1983) The relationship between the Rht1 and Rht2 dwarfing genes and grain weight in Triticum aestivum L. spring wheat. Theor Appl Genet 66:153–157. https://doi.org/10.1007/BF00265191
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis 84:203. https://doi.org/10.1094/PDIS.2000.84.2.203B
Qureshi N, Kandiah P, Gessese MK, Nsabiyera V, Wells V, Babu P, Wong D, Hayden M, Bariana H, Bansal U (2018) Development of co-dominant KASP markers co-segregating with Ug99 effective stem rust resistance gene Sr26 in wheat. Mol Breed 38:97. https://doi.org/10.1007/s11032-018-0854-6
Ramirez-Gonzalez RH, Segovia V, Bird N, Fenwick P, Holdgate S, Berry S, Jack P, Caccamo M, Uauy C (2015) RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol J 13:613–624. https://doi.org/10.1111/pbi.12281
Rasheed A, Wen W, Gao F, Zhai S, Jin H, Liu J, Guo Q, Zhang Y, Dreisigacker S, Xia X, He Z (2016) Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet 129:1843–1860. https://doi.org/10.1007/s00122-016-2743-x
Semagn K, Babu R, Hearne S, Olsen M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14. https://doi.org/10.1007/s11032-013-9917-x
Shorinola O, Balcárková B, Hyles J, Tibbits JFG, Hayden MJ, Holušova K, Valárik M, Distelfeld A, Torada A, Barrero JM, Uauy C (2017) Haplotype analysis of the pre-harvest sprouting resistance locus Phs-A1 reveals a causal role of TaMKK3-A in global germplasm. Front Plant Sci 8:1555–1555. https://doi.org/10.3389/fpls.2017.01555
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481. https://doi.org/10.1146/annurev-phyto-072910-095423
Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MA, Bhavani S, Rouse MN, Pretorius ZA, Szabo LJ, Huerta-Espino J, Basnet BR, Lan C, Hovmøller MS (2015) Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105:872–884. https://doi.org/10.1094/PHYTO-01-15-0030-FI
Srinivasachary GN, Steed A et al (2008) Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor Appl Genet 118:695–702. https://doi.org/10.1007/s00122-008-0930-0
Sukumaran S, Lopes M, Dreisigacker S, Reynolds M (2018) Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet 131:985–998. https://doi.org/10.1007/s00122-017-3037-7
Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y (2016) A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol CB 26:782–787. https://doi.org/10.1016/j.cub.2016.01.063
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301. https://doi.org/10.1126/science.1133649
Vetch JM, Stougaard RN, Martin JM, Giroux MJ (2019) Review: revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.). Plant Sci 281:180–185. https://doi.org/10.1016/j.plantsci.2019.01.004
Wang S, Yan X, Wang Y et al (2016) Haplotypes of the TaGS5-A1 gene are associated with thousand-kernel weight in Chinese bread wheat. Front Plant Sci 7:783. https://doi.org/10.3389/fpls.2016.00783
Wilhelm EP, Mackay IJ, Saville RJ, Korolev AV, Balfourier F, Greenland AJ, Boulton MI, Powell W (2013) Haplotype dictionary for the Rht-1 loci in wheat. Theor Appl Genet 126:1733–1747. https://doi.org/10.1007/s00122-013-2088-7
Wilkinson PA, Winfield MO, Barker GL et al (2012) CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinformatics 13:219. https://doi.org/10.1186/1471-2105-13-219
Zhang L, Zhao Y-L, Gao L-F, Zhao GY, Zhou RH, Zhang BS, Jia JZ (2012) TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol 195:574–584. https://doi.org/10.1111/j.1469-8137.2012.04194.x
Zhao J, Wang Z, Liu H, Zhao J, Li T, Hou J, Zhang X, Hao C (2019) Global status of 47 major wheat loci controlling yield, quality, adaptation and stress resistance selected over the last century. BMC Plant Biol 19:5. https://doi.org/10.1186/s12870-018-1612-y
Zheng J, Liu H, Wang Y, Wang L, Chang X, Jing R, Hao C, Zhang X (2014) TEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L.). J Exp Bot 65:5351–5365. https://doi.org/10.1093/jxb/eru306
Zikhali M, Wingen LU, Griffiths S (2016) Delimitation of the earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J Exp Bot 67:287–299. https://doi.org/10.1093/jxb/erv458
Funding
This work was supported by the BecA-ILRI Hub’s Africa Bioscience Challenge Funds Fellowships to MW and ZT, the Royal Society and African Academy of Sciences FLAIR fellowship to OS, and the UK Biotechnology and Biological Sciences Research Council (BBSRC).
Author information
Authors and Affiliations
Contributions
OS, CU, NY, and RW designed the experiments. MW, ZT, LM, HZ, MR, and OS performed the experiments (sampling, genotyping, and field work) and analyzed the results. MW, ZT, and OS wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mercy Wamalwa and Zerihun Tadesse co-first authors
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wamalwa, M., Tadesse, Z., Muthui, L. et al. Allelic diversity study of functional genes in East Africa bread wheat highlights opportunities for genetic improvement. Mol Breeding 40, 104 (2020). https://doi.org/10.1007/s11032-020-01185-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-020-01185-x