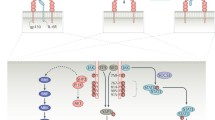
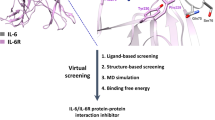
Abstract
IL(interleukin)-6 is a multifunctional cytokine crucial for immunological, hematopoiesis, inflammation, and bone metabolism. Strikingly, IL-6 has been shown to significantly contribute to the initiation of cytokine storm—an acute systemic inflammatory syndrome in Covid-19 patients. Recent study has showed that blocking the IL-6 signaling pathway with an anti-IL-6 receptor monoclonal antibody (mAb) can reduce the severity of COVID-19 symptoms and enhance patient survival. However, the mAb has several drawbacks, such as high cost, potential immunogenicity, and invasive administration due to the large-molecule protein product. Instead, these issues could be mitigated using small molecule IL-6 inhibitors, but none are currently available. This study aimed to discover IL-6 inhibitors based on the PPI with a novel camelid Fab fragment, namely 68F2, in a crystal protein complex structure (PDB ID: 4ZS7). The pharmacophore models and molecular docking were used to screen compounds from DrugBank databases. The oral bioavailability of the top 24 ligands from the screening was predicted by the SwissAMDE tool. Subsequently, the selected molecules from docking and MD simulation illustrated a promising binding affinity in the formation of stable complexes at the active binding pocket of IL-6. Binding energies using the MM-PBSA technique were applied to the top 4 hit compounds. The result indicated that DB08402 and DB12903 could form strong interactions and build stable protein–ligand complexes with IL-6. These potential compounds may serve as a basis for further developing small molecule IL-6 inhibitors in the future.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The SARS-CoV-2 pandemic, nicknamed COVID-19, has emerged as a severe public health challenge. One of the dangerous complications of COVID-19 disease is the overwhelming release of proinflammatory cytokines, which in turn trigger uncontrolled inflammatory responses known as “cytokine storms” [1, 2]. Cytokine storms can rapidly cause acute respiratory distress syndrome (ARDS), multiple organ failure, and poor prognosis in COVID-19 patients [3]. Recent studies of the immunological profiles of critically ill patients with Covid-19 have shown overactivity of the humoral immune pathway with interleukin IL-6 amplification. This results in the overproduction of inflammatory cytokines and chemokines, notably IL-6, ultimately resulting in a cytokine storm with ARDS [4]. Furthermore, according to analyses of serum IL-6 levels in covid19 patients, the results showed that patients with complicated forms of COVID-19 had nearly three-fold higher levels of IL-6 compared with uncomplicated patients [5]. Therefore, IL-6 inhibition could become a novel target for therapeutics to manage dysregulation in COVID-19 patients, and studies aimed at interfering with this interleukin activity are urgently required.
IL-6 was identified in 1986 as a B cell stimulation factor that enhances the differentiation of effector B cells toward antibody-producing cells, also known as IFN-β2 [6, 7]. IL-6 is a 185 amino acid polypeptide of 22 kDa, derived from a wide variety of immune and non-immune cells, including T cells, B cells, monocytes, endothelial cells, fibroblasts, keratinocytes, mesangial cells, and adipocytes [7, 8]. The IL-6 type cytokines are activated through the gp130 signaling probe, a transmembrane protein found in most cytokine receptor systems to activate the Janus family of tyrosine kinases (JAKs), including JAK1, JAK2, and Tyk2 [9, 10]. The connection between IL-6 and gp130 is formed through an IL-6-specific receptor (IL-6R or gp80) [11]. On target cells, the extracellular domains of IL-6R and gp130 associate with IL-6 to create a functional hexameric signaling complex, which then binds to JAK proteins and initiates downstream signaling pathways [12, 13]. IL-6 has two modes of action: cis and trans. Classic (cis)-signaling occurs only in cells expressing membrane-bound IL-6 receptor (mIL-6R), which has homeostatic and anti-inflammatory effects. On the other hand, in IL6 trans-signalling, IL6 binds to a soluble IL6R (sIL6R) and possesses detrimental pro-inflammatory effects [13].
Thus far, IL-6 pathway inhibitors such as tocilizumab, siltuximab, and sarilumab have been developed and are used to treat several chronic inflammatory diseases, including rheumatoid arthritis (RA) [14, 15]. Significantly, these targeted monoclonal antibodies (mAb) have also been shown to improve survival in patients with severe coronavirus disease 2019 (COVID-19) [15, 16]. Moreover, according to recent studies, blockade of IL-6 signaling by directly blocking the interaction between IL-6 and IL-6R also reduces the progression of cytokine storm [16, 17]. Although the benefits obtained from monoclonal antibody treatment have been demonstrated, these biological agents still have certain disadvantages, such as high treatment cost, unsuitability for oral administration, difficulty to control, and the risk of inducing strong immune responses. Small molecule therapy, on the other hand, provides a number of significant advantages, including simpler oral administration, possible optimization of pharmacokinetic properties, lower treatment costs, more controllability, and less immunogenic [18]. Oral bioavailability offers significant benefits in terms of accessibility, compliance, and widespread use, making such therapy more suitable for long-term treatment.
The present study was designed to identify potential compounds as IL-6 inhibitors by computational methods. The strategy includes three main stages. Firstly, we generated structure-based pharmacophore models for IL-6 and used them for the initial screening of the Drugbank database. Secondly, molecular docking is performed to determine the suitability of the small molecule in the binding site. Finally, top compounds were validated by ADME analysis, molecular dynamics (MD) simulations, and free binding energy. As to structure-based drug design, molecular docking offers a starting model of the binding modes between small molecules with protein at the atomic level. MD simulations are subsequently used to evuluate the stability of these complexes in the context of the full atomistic molecular mechanics force field [19].
Materials and methods
Drugbank databases set for virtual screening
The databases used for screening included 9,213 compounds from the DrugBank database [20]. The next step was conformationally prepared using the conformational Import tool in Molecular Operating Environment (MOE) 2015.10 software [21]. The conformation limit was set to 10,000, the stochastic search iteration limit was 1,000, the energy minimization iteration limit was 1,000, the energy minimization gradient test was 0.0001, and the input filters were clear. The output conformers were saved as MOE database (*.mdb).
Pharmacophore model construction
The pharmacophore models were constructed based on a protein–protein interaction at the binding site 1 of interleukin 6 with 68F2 that mimics the interaction of IL-6 with IL-6R [9]. According to X-ray structure analysis, the side chain of HCDR3 valine ties into site 1 like IL-6R Phe279, while the side chain of LCDR1 tyrosine occupies a second cavity within site 1 and mimics the interactions of IL-6R Phe229. Phe229 and Phe279 of IL-6R are called the “hot spot residue” by Boulanger et al. [12] because mutagenesis studies confirmed their critical role in the interaction between the receptor and the cytokine. Mutation of this residue to valine or serine completely abolishes the IL-6R binding to IL-6 [22]. The interaction between Phe229 and Phe279 from IL-6R and the cytokine is mimicked by two hydrophobic residues from the CDR loops of 68F2. Here, the hydrophobic side chain of Tyr32 located in the CDR1 loop of the variable domain light chain superimposes Phe229. Similarly, the side chain of Val104 of the CDR3 loop of the heavy chain takes over the role of Phe279. The full set of interactions (hydrogen bonds, salt bridges, and hydrophobic) was observed between antibody 68F2 and antigen IL-6 in the X-ray structure. Therefore, the 68F2 antibody can be seen as an accurate structural mimic of IL-6R: it occupies the same interaction site with and provides similar key interacting residues to anchor points on the IL-6 surface [9]. Protein contact analysis between IL-6 and 68F2 by MOE 2015 software showed that Tyr32 and Val104 of 68F2 interact mainly with amino acids Phe74, Gln75, Leu178, and Arg179 of IL-6. Previous studies on hot-spot residues mutations also indicated that Arg179, Leu178, and Phe74 are involved in the interaction between IL-6/IL-6R [22]. Consequently, in this study, the 3D-pharmacophore model is constructed based on the chemical features of these residues and the spatially favorable locations of their functional groups. This step was performed by the pharmacophore editor tool in the MOE 2015.10 software [21]. The interactions of 4 amino acids Phe74, Gln75, Leu178, and Arg179 on IL-6 are specified in Table 1.
Molecular docking
The protein structures were prepared (high-resolution crystal structures 2.9Å, PDB ID: 4ZS7) [23], including the repair of amino acid residues, the addition of hydrogen atoms and charges, the removal of water molecules, and the minimization of energy. The ligand databases were minimized the energy using MOE 2015.10 software with Amber10: EHT force field [21]. The results were converted and saved as a *.sdf format suitable for the docking software.
The protein-prepared structure above was fed into the BioSolveIT LeadIT 2.1.8 software [24] to create the binding site. The crucial residues were used as references to generate the binding site model. They were loaded into the docking program, and a binding pocket was generated by being expanded with a suitable radius. Docking runs were operated using the following parameters: the maximum number of solutions per iteration was 1000, that per fragmentation was 200, the number of poses to keep was the top 10, and default docking options were used [25]. Successfully docked ligand conformations were evaluated on the lowest binding energy (kJ/mol). The frequency interaction between ligands with the critical residues of IL-6 was analyzed using the protein–ligand interaction fingerprints (PLIF) tool of the MOE 2015.10 software. PLIF summarizes the interactions between ligands and proteins using a fingerprint scheme. The interactions are classified into six types: side chain hydrogen bond donor (D), side‐chain hydrogen bond acceptor (A), backbone hydrogen bond donor (d), backbone hydrogen bond acceptor (a), ionic attraction (I), arene–arene interaction (R), and surface contact (C) [26].
Analysis of ADME properties
SwissADME online free tool was employed to evaluate the individual ADME behavior like physiochemical properties, lipophilicity, water solubility, pharmacokinetics, drug-likeness, and medicinal chemistry properties of the compounds. The best ligands after molecular docking were uploaded directly to the submission page (http://www.swissadme.ch) in *.smlie format, and the results are presented for each molecule in tables, graphs, and an excel spreadsheet. The physicochemical parameters were generated, including the molecular weight between 150 and 500 g/mol, polarity: TPSA between 20 and 130 Å2, solubility: log S not higher than 6, and flexibility: no more than ten rotatable bonds. For lipophilicity, we used the descriptor Consensus LogP(o/w), the arithmetic means of the logP(o/w) values predicted by the five proposed methods. The solubility of substances is better when LogP is less than 5. The water solubility of compounds was predicted based on the ESOL LogS values, where logS should be from − 6.5 to 0.5 mol/dm3 [27]. The BOILED-Egg allows for the intuitive evaluation of passive gastrointestinal absorption (HIA) and brain penetration (BBB) in the function of the position of the molecules in the WLOGP- versus -TPSA referential [28,29,30].
Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out using GROMACS 2021.2 software [31]. Protein topologies were prepared by the pdb2gmx module of GROMACSusing the CHARMM27 all-atom force field [32]. The simulations were performed on IL-6, in the apoprotein form and complexes with small molecules. Ligand topologies were constructed by the Swissparam web server [33] after adding hydrogen atoms to the initial structure by the Avogadro software [34]. A dodecahedron box was defined where the protein was positioned at least 10 Å from the box edge. The system was filled with water solvent (TIP3P model) and electrically neutralized by the addition of an appropriate number of Na+ or Cl− ions (salt concentration was 0.15 M) [35]. The constructed system was energy minimized for 100 ps using the steepest descent minimization with a maximum force of 10 kJ/mol. Production runs of 100 ns were performed at 300 K for the NVT (isothermal-isochoric) ensemble and 1.0 bar pressure for the subsequent NPT (isothermal-isobaric) ensemble. After the MD production had been completed, data from the resulting trajectories were used to calculate the root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and solvent accessible surface area (SASA) values by GROMACS built-in commands to evaluate the stability of the complexes and the effect of ligands on the receptor when binding occurs. In particular, the occupancy of hydrogen bond formation was also analyzed using the VMD software [36] to determine the interacting ability of ligands with the key residues. A hydrogen bond was defined by simple geometric criteria: a distance between hydrogen donor (D) and acceptor (A) atoms of < 3.5 Å and an angle of D–H…A > 120° [37].
Binding free energy
The molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) method was used to compute the binding free energy of all complexes. A thousand frames were extracted from each 100 ns simulation trajectory by means of the gmx-trjconv utility of Gromacs. The extracted frames were utilized for free binding energy calculation in the company with the g_mmpbsa tool [38]. The total binding free energy (∆Gbind) was calculated as:
The free energy of protein, ligand, or complex component can be calculated as follows:
where EMM is the molecular potential energy between a ligand and the biomolecular target and is contributed by binding energy (Ebond) and unbinding energy (Evdw + Eelec). Ebond is the energy of bonded interactions which is calculated as zero in a dynamic simulation, Evdw and Eelec are the van der Waals and the electrostatic interactions energy, respectively. Gsolvat is calculated from polar and non-polar components of molecules, Gpolar represents the polar contributions between the solute and solvent to the solvation energy, and GSASA is the non-polar solvation energy using the solvent accessible surface area (SASA). In the MM/PBSA approach, the entropy contribution can be omitted if relative binding free energies of different ligands for the same protein shall be computed [39,40,41,42]. Hence, the binding free energy was calculated as the sum of van der Waals energy, electrostatic energy, polar solvation energy, and solvent-accessible surface area energy (SASA) [38], and the formula can be rewritten as:
Results and discussions
Pharmacophore model generation
Using a manually created pharmacophore query at the binding site, two 3D-pharmacophore models were generated based on the PPIs of IL-6/68F2. Ph-IL6-1 is the first pharmacophore model, which was constructed from the residues of the 68F2 antibody corresponding to the crucial residues of IL-6, including Tyr32 and Val104. The fundamental structural properties of two residues were selected to create five pharmacophore features by the Pharmacophore editor tool of MOE version 2015.10. In detail, the used three functional groups of Tyr32, including a phenyl ring that is prone to participate in aromatic interaction (Aro), a donor hydrogen bond group (Don)–amino (–NH2), and a carboxylate group (–COO−) that can accept hydrogen bond (Acc). Using a similar approach to Val104, another two pharmacophore points were built, and the final developed pharmacophore contained F1: Aro, F2: Don, F3: Acc, F4: Hyd, and F5: Ani (Fig. 1A).
The second pharmacophore model (Ph-IL6-2) contained five features that must complement functional groups of IL-6, covering Phe74, Gln75, Leu178, Arg179, and Gln183 of IL-6. As previously known, the -N+ ion in Arg179 is considered by default as a cation (Cat), and a pharmacophore feature ani (anion) will be placed on the projection of the nitrogen atom. In that way, the other pharmacophore feature was generated as follows: F1: Aro, F2: Don, F3: Acc, F4: Hyd, and F5: Ani. Because Phe74, Leu178, and Arg179 are crucial residues, corresponding F1, F4, and F5 were essential points, and either F2 or F3 was constrained at least one point in this model (Fig. 1B).
A database of more than 5 million conformations of the Drugbank library was rapid and virtually screened in turn through the two pharmacophores above. The screening resulted in 1,426 compounds satisfying both models, of which 1174 satisfied the Ph-IL6-1 model and 252 satisfied the Ph-IL6-2 model. All compounds that met the two models were further screened by molecular docking as the second filter of the drug discovery process.
Molecular docking and virtual screening
Molecular docking is one of the most frequently used methods in structure-based drug design because of its ability to predict, with a remarkable degree of accuracy, the conformation of small molecule ligands was placed into the appropriate target binding site. Therefore, a molecular binding simulation was performed in this study to elucidate the binding mode of the ligand-receptor. Accordingly, a binding pocket with a radius of 10 Å was extended from the locations of key residues Phe74, Gln75, Leu178, Arg179, and Gln183. The generated molecular docking model is illustrated in Fig. 2A.
The compounds obtained from screening through the pharmacophore models were used for docking into the binding pocket of IL-6. As a result, 1364 compounds (95.6%) successfully docked to IL-6 with negative docking scores; 1.8%, 76.9%, and 21.3% of which had docking scores < − 20 kJ/mol, from − 20 to − 10 kJ/mol and from − 10 to 0 kJ/mol, respectively (Table S1 and Figure S1). A more negative docking score indicates stronger binding of the ligand to the protein target. The top 24 ligands with docking scores less than -20 kJ/mol and interacting with at least three critical residues at the determined binding pocket on IL-6 are shown in Fig. 2B.
From the docking results, we further applied the Protein–ligand interaction fingerprints (PLIF) in the MOE to analyze the presence of different types of chemical interactions between ligands and the protein target. This technique also aims to evaluate and improve the performance of the structure-based 3D-pharmacophore models in virtual screening. The frequency of ligands interacting with binding site residues was counted and illustrated in Fig. 3, Figure S2. Based on the PLIF results, it can be seen that a significant proportion of ligands could interact with the key residues. In particular, Phe74, Gln75, Arg179, and Gln183 of IL-6 formed interaction with ligands at the high ratio of 88%, 87%, 76%, and 70%, respectively. On the other hand, from docking models of good bonding, the ligands were required as follows: the hydrogen bond donor and acceptors that can combine with the polar amino acids (Gln75, Gln183); the hydrophobic group that can go deep into the binding site to interact with Leu178; the ions which will create the salt bridge bonding with the charged amino acid (Arg179, Arg182), and the aromatic rings can form arene-arene interactions with Phe74 (Table S2).
In general, the PLIF results increased the reliability and screening efficiency of 3D-pharmacophore by identifying compounds with suitable physicochemical properties for binding to IL-6 from databases as diverse as drug banks. Based on the docking score and interaction analysis, the top 24 ligands were selected for further screening steps, and their detailed interaction models are shown in Table 2 and Table S3.
ADME analysis
A major crucial factor determining the success of any oral drug discovery and development is the prediction of ADME parameters before switching to experimental studies. Therefore, the 24 top docking score compounds were further tested for their ADME parameters using the SwissADME tool, a freely available web tool for pharmacokinetic predictions, drug-likeness, and suitable for the medicinal synthesis of these small molecules [28]. Compounds with pharmacokinetic parameters that do not meet “drug-like” properties will be rejected in the next step of the screening process.
Table 3 and Figure S3 showed physicochemical and bioavailability radar for the top list (24 ligands). Most of these compounds had molecular weight varied between 150 and 500 g/mol, which is a prime property to be called drug-likeness of the small molecules. The number of rotatable bonds can predict molecular flexibility and permeability. The molecular permeability is strongly correlated with the number of rotatable bonds without considering the molecular weight. Compounds with more than ten rotating bonds have been shown to have poor permeability. Interestingly, all the top compounds had a number of rotatable bonds of less than 10 that have good oral absorption. The numbers of H-bond acceptors and donors were in the range of 3–10 and 1–4, respectively. Table 3 and Figure S3 also described these top lists’ ESOL LogS values and solubility categories, the results showed that all compounds are soluble in water. Among them, DB12008 is the best water-soluble compound with a minimum logS value of -2.89 and a maximum logS value of -6.06, DB08881 is considered the least water-soluble compound.
BOILED-Egg is an intuitive method for simultaneously predicting two key ADME parameters, including passive gastrointestinal absorptive capacity (HIA) and brain accessibility (BBB) via the WLOGP and TPSA reference values. In the BOILED-Egg plot (Fig. 4), the yellow region characterizes the boundary of molecules’ properties related to passing the blood–brain barrier (BBB), and the white region defines the properties of molecules with a highly probable HIA absorption. Meanwhile, the outer space gray is associated with poor human intestinal absorption (HIA) and limited brain penetration. As observed from the graph, with the exception of DB08881 and DB11772, it can be seen that most of the top compounds are located inside the egg—an area with suitable physicochemical space for oral bioavailability. Notably, for DB06190 and DB12658, even though these two substances are in the yellow region, they are P-glycoprotein (PGP) substrates, which seem to be readily released from the central nervous system (CNS). Additionally, in the graph, the blue dots are for the P-gp (PGP+) substrate, which can be actively effluxed from the brain or gastrointestinal tract, while the red dots are for P-gp non-substrate (PGP˗). Based on the finding, more than 60% of the candidates were PGP+, suggesting that they may have reduced intestinal absorption but exerted a CNS protective effect.
Drug metabolism and elimination are profoundly influenced by the cytochrome P450, including CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. Inhibition of five major isoenzymes is one of the main causes of toxicity or other undesirable effects. These pharmacokinetic disadvantages are undoubtedly due to the lower clearance and accumulation of the drug or its metabolites. Table S4 provides data on whether selective inhibition of CYP isozymes by top virtual screening hits. Unfortunately, all 24 ligands interact with at least one of the five metabolic enzymes above. Due to the inhibition of all five isozymes above, DB11830, DB12658, and DB08911 are predicted to have greater drug-drug interactions than others.
Lipinski’s rule of five [43] is commonly applied to evaluate the similar ‘drug-like’ properties of small molecules. The biologically active molecule must meet five criteria to be potentially used as a drug for oral administration: MW ≤ 500, MLOGP ≤ , N or O ≤ 10, and NH or OH ≤ 5. Intriguingly, with the exception of DB11772, DB08911, DB03115, and DB13060 (MW > 500), the remaining 20 ligands all follow Lipinski's rule. Besides, this top list of compounds also agreed with other models of drug-likeness, namely Ghose [44], Veber [45], Egan [46], and Muegge [47] (Table 5).
When we evaluated AMDE profiles of the top 24 hits from the virtual screening, seven compounds stood out: DB07970, DB08402, DB11963, DB12903, DB07101, DB07586, and DB07268. These molecules have physicochemical properties suitable for oral bioavailability, are predicted to be non-substrate of P-gp (PGP˗) and do not cross the BBB. Therefore, the 100 ns MD simulation was performed for seven protein–ligand complexes to validate the interactions of the candidate molecules with the IL-6.
Molecular dynamics simulation
MD simulation studies play a significant role in the drug discovery process. In this study, structural changes, stability, and flexibility of the IL-6 protein to the ligands were monitored by comparing the values of root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent accessible surface areas (SASA), a radius of gyration (Rg) and the occupancy interaction of hydrogen bonds (H-bonds).
RMSD evaluates the structural stability of ligand–protein complexity over a specific time period. The RMSD values of individual protein-IL6 (apoprotein) and the complexes (docked forms) are shown in Fig. 5A and Table 4. According to the results in Table 4, out of the seven molecules studied, four molecules (DB07970, DB08402, DB11963, and DB12903) have RMSD values lower than that of the apoprotein (3.26 ± 0.47 Å). For all compounds, DB07101 has the highest RMSD of all systems with a mean value of 4.14 ± 0.43 Å, and this value tends to increase as the simulation run gets longer. It indicates that the DB07101 complex had less stability during the simulation. Meanwhile, DB08402 achieves the lowest RMSD with a mean value of 2.84 ± 0.24, and the complex of IL-6 with DB08402 was predicted to have the most stability during the above 100 ns simulation. From the RMSD plots in Fig. 5A, we can see that the RMSD of the apoprotein-IL6 strongly deviates in the initial 30 ns, but they become stable after that till the end of the simulation time. Similarly, DB07970 and DB11963 in their complexes with IL-6 also fluctuated strongly during the first 30 ns of the simulation progress. Still, after fluctuation time, these complexes gradually reached equilibrium and had smaller RMSD fluctuations (< 1 Å) than apoprotein (Fig. 5A).
Solvent accessible surface area (SASA) of proteins has always been considered a decisive factor in protein folding and stability studies. A higher SASA value indicates structural relaxations of protein, and a low fluctuation is expected over the simulation time [48]. The average values of all systems are shown in Table 4, and a graph showing their change over time is shown in Figure S4. Except for DB07101 and DB07268, which shows higher SASA values than apoprotein (85.73 ± 0.90 nm2) and reveal the conformational states with higher protein expansion, the protein structure in all complexes with the remaining ligands is virtually unaffected by ligands binding.
The compactness of a system is measured by the radius of gyration (Rg). According to Lobanov et al. (2008), Rg is considered an indicator of the folding rate and the compactness of a protein structure [49]. A more compact protein indicates that the drug molecule has not significantly interfered with the folding mechanism of the protein. Figure 5S describes the radius of the gyration plot for IL-6 in docked and undocked form, their mean values are shown in Table 4. Interestingly, all of the docked forms with seven ligands have Rg values of approximately or less low than apporotein (12.18 ± 0.16 Å). These results clearly indicate that the protein structure is not significantly affected by the binding of the ligands.
The RMSD of the ligand also determines the protein flexibility, if the molecule fluctuations from the protein site, the molecule may lose binding and the stability of the complex will be reduced. The average ligand RMSD values are in Table 4 and the RMSD-time plot is in Fig. 5C. The results show that most complexes had RMSD values smaller than 2 Å. Strikingly, DB08402, DB07586, and DB12903 reached stability with RMSD fluctuations only about 1 Å after 5, 10, and 20 ns, respectively. Thus, we can consider that all complexes keep a steady stage throughout the simulation.
To provide more detailed information on motions in IL-6 binding of selected compounds. RMSF analysis was performed to reveal information on residue-specific flexibility. A more stable structure has a low RMSF value, while a higher RMSF value represents a higher degree of movement [50]. The RMSF profiles of the apoprotein and the complexes calculated by residues index Cα overlapped with that of the apoprotein as shown in Fig. 5B.
As observed in the RMSF plots, protein fluctuates strongly mainly in 2 regions: Region1-residues 40 to 70, which majorly belong to the interpolated flexible loops of the inter-helix between helices A and B, and Region2-residues 130 to 160, which are lying outside the principal four helical bundles and is also flexible in nature. Interestingly, the critical residues at the binding site, such as Phe74, Gln75, Leu178, Arg179, and Gln183 of IL-6 had stable fluctuations with RMSF < 2.0 Å in all systems. The results strongly suggested that all the ligand-bound IL-6 complexes were stable.
The number of intermolecular hydrogen bonds was analyzed to get insight into the protein–ligand interaction and stability. In this study, the number of hydrogen bonds was calculated from the trajectories of 100 ns MD simulations using the VDM software. Table S6 and Fig. 5D indicate that all ligands interacted strongly with IL-6. At the same time, the percentage occupancy of hydrogen bonds of these ligands was also investigated to find compounds that interact strongly with the critical residues of IL-6/68F2 PPIs. Excepted for DB07268 and DB07586, the remaining five ligands with the H-bond occupation of > 100% were considered strong hydrogen bonds (Table S6).
By comparing the values of protein backbone RMSD, solvent accessible surface area (SASA), radius of gyration (Rg), ligands RMSD, RMSF, and the hydrogen bond interaction with key residues, the four ligands DB12903, DB11963, DB07970, and DB08402 were identified as the most potent inhibitors of the IL-6.
In addition to hydrogen bonding, the PPIs of IL-6/68F2 are formed mainly by hydrophobic interactions with hot-spot radicals. Therefore, we further analyzed the surface contact and arene-arene interactions between IL-6 and the top 4 ligands. The results were extracted from 100 frames of 100 ns MD simulation trajectories using the PLIF tool in MOE 2015.10 software and are detailed in Table S7. Analysis of interaction occupancy on each ligand binding to key residues of IL-6 showed that all four ligands exhibited strong interactions with the key residues. Some residues had a total interaction frequency > 100% because they formed multiple same interactions simultaneously.
For more detail, visual analyses were conducted by comparing the poses for each ligand-target interaction in the protein active sites. First, we discuss the analysis for DB08402, in which its interaction is shown in Fig. 6D, Tables S6, and S7. The binding mode of DB08402 indicates the importance of hydrogen bonds and hydrophobic interactions within the activity site of IL-6. This ligand form strong interactions with the two hot-spot residues of IL-6, including Arg179 and Phe 74. In detail, the oxygen atom of the carboxylate group (–COO−) at the benzoate ring and the carbonyl group (–CO−) of the 2,4 dichlorobenzamido ring of DB08402 accept the hydrogen bonding of Phe74 and Arg179 with a high-frequency of 156.28% and 95.18%, respectively. For the hydrophobic interaction, the aromatic rings of DB08402 formed edge‐to‐face contact with Phe74 by Pi-alkyl and arene-arene stacking with a total frequency of 88.11% and 31.19%, respectively. In continuation, these aromatic rings of DB08402 also form Pi-cation interactions with the N atoms (–N+), Pi-alkyl interactions with the carbon backbone, and alkyl interactions with chlorine atoms of Arg179 with a total frequency of 72.92%.
Next, we discuss the DB12903 and DB11963 target interactions, which are presented in Fig. 6A, Fig. 6B, Table S6, and Table S7. The results show that DB12903 and DB11963 have the common property that both create a strong hydrogen binding with Arg179, Gln 183, and Gln75. The ˗NH+ and ˗NH2 groups of these ligands donated hydrogen bonds with Gln75 with a frequency of 69.41% and 121.76%, respectively. Similarly, Gln183 has a percentage occupancy of 93.19% and 120.01%, respectively. On the other hand, these two ligands act as hydrogen acceptors from the amino groups of Arg179 with percent of occurrence values of 170.04% and 205.70%, respectively. For hydrophobic interaction, both ligands also show a high occupancy interaction with Gln75 and Arg179. In the case of DB12903, the indol, pyrazole, and benzothiazole rings of DB12903 interact with the backbone atoms of Arg179 and Gln175 with a frequency of 97.14% and 79.04%, respectively. In this orientation, the quinazoline and phenyl rings of DB11963 also contact with the two residues above with an occupancy of 96.97% and 90.02%, respectively.
Finally, we discuss the DB07970-target interaction shown in Fig. 6C, TableS6, and Table S7. The results show that DB07970 has high-frequency hydrogen bonds and hydrophobic interactions with Arg179 residue. Here, the nitrogen, oxygen, and fluoride atoms of DB07970 accepted hydrogen bonds from Arg179 with a frequency of 168.94%, and the aromatic rings participated in Pi-cation and Pi-alkyl interactions with these hot-spot residues with a total percent of occurrence value of 89.25%.
To take a closer look at the binding strength between the ligand and the protein target, we extract frames at 0 ns, 50 ns, and 100 ns time points, respectively. As expected, all four ligands, DB12903, DB11963, DB07970, and DB08402, could maintain contact with IL-6 during 100 ns MD simulation (Fig. 7).
At the end of the MD simulation, the binding free energy of inhibitors was calculated by MM/PBSA approach. Data derived from the average binding energy calculations can offer a better insight into the interactions of the enzyme-ligand complexes. MM/PBSA was employed on the MD trajectories based on van der Waals, electrostatic, polar solvation, and nonpolar solvation energies (SASA). Based on these results in Table 5, the binding free energy of DB12903, DB11963, DB07970, and DB08402 are − 118.30 ± 23.88 kJ/mol, − 91.70 ± 34.80 kJ/mol, − 129.96 ± 39.31 kJ/mol, and − 267.09 ± 40.96 kJ/mol, respectively. This observation highlighted that DB08402 forms a powerful interaction with IL-6 and secures the top rank in the energy list among selected compounds. Followed by DB07970 and DB12903 with 2nd and 3rd place, respectively. Finally, with the highest ∆Gbind values, DB11963 is determined as the ligand with the lowest binding affinity.
However, when observing the fluctuations of the complexes, the results show that DB08402 and DB12903 complexes give a stable energy value, whereas that of DB07970 and DB11963 fluctuate enormously and had positive ∆Gbind values at the last 20 ns of MD trajectory (Fig. 8). Notably, DB08402 binding energy tended to decrease at the end of the simulation and reflected an increase in binding affinity.
In summary, based on analysis of complexes stability, frequency of interactions occupancy, and MM/PBSA binding free energy, DB08402, and DB12903 were identified as the best-hit compound within the scope of this study. The results of virtual screening, PLIF analysis, ADME properties prediction, and MD simulations are summarized in Fig. 9.
Conclusions
As we know, protein–protein interactions (PPIs) could be a target for blocking signal transduction. In recent years, many new drug discovery projects have been proposed to develop novel PPI inhibitors to find potential therapy in related diseases, even for COVID-19 [51,52,53,54]. However, because the binding pocket's shape has not been well described on the protein surface, it is challenging to discover small molecules with PPI inhibitory potential. Fortunately, the advent of X-ray crystallography and mutation studies have contributed to elucidating the nature of PPI between IL-6 and its receptor IL-6R. In addition, the significant development of artificial intelligence (AI) in recent decades has significantly reduced the time and cost of new drug discovery [55]. From there, the medicinal chemist can begin research on IL-6 inhibitors based on these fundamental biological databases.
In this study, we approach the drug discovery process by in silico screening. With the ability to quickly search for compounds from large databases, this method will enhance the hit rate and reduce the time and cost of in vitro and in vivo testing. The process was constructed and screened from the approximately 10.000 substances in the Drugbank database through structure-based pharmacophore and molecular docking models. 24 compounds with docking scores < –20 kJ/mol interacted with critical residues of IL-6 were identified. Subsequently, applying the SwissADME tool resulted in seven druggable candidates with suitable pharmacokinetic properties. Calculations based on 100 ns MD simulation data and free binding energy indicated DB12903 and DB08402 as promising the most potential IL-6 inhibitors candidate. Hopefully, the outcomes of this research will provide a novel therapeutic approach for inflammatory, autoimmune, and infectious diseases, especially for COVID-19.
References
Jiang Y, Rubin L, Peng T, Liu L, Xing X, Lazarovici P, Zheng W (2022) Cytokine storm in COVID-19: from viral infection to immune responses, diagnosis and therapy. Int J Biol Sci 18:459–472. https://doi.org/10.7150/ijbs.59272
Ogata A, Kato Y, Higa S, Yoshizaki K (2019) IL-6 inhibitor for the treatment of rheumatoid arthritis: a comprehensive review. Mod Rheumatol 29:258–267. https://doi.org/10.1080/14397595.2018.1546357
Gorham J, Moreau A, Corazza F, Peluso L, Ponthieux F, Talamonti M, Izzi A, Nagant C, Djangang NN, Garufi A, Creteur J, Taccone FS (2020) Interleukine-6 in critically ill COVID-19 patients: a retrospective analysis. PLoS ONE 15:e0244628. https://doi.org/10.1371/journal.pone.0244628
Abdin SM, Elgendy SM, Alyammahi SK, Alhamad DW, Omar HA (2020) Tackling the cytokine storm in COVID-19, challenges and hopes. Life Sci 257:118054. https://doi.org/10.1016/j.lfs.2020.118054
Coomes EA, Haghbayan H (2020) Interleukin-6 in Covid-19: a systematic review and meta-analysis. Rev Med Virol 30:1–9. https://doi.org/10.1002/rmv.2141
Kishimoto T (2010) IL-6: from its discovery to clinical applications. Int Immunol 22:347–352. https://doi.org/10.1093/intimm/dxq030
Scheller J, Chalaris A, Schmidt-Arras D, Rose-Jone S (2011) The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 1813:878–888. https://doi.org/10.1016/j.bbamcr.2011.01.034
Somers W, Stahl M, Seehra JS (1997) 1.9 Å crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. Embo J 16:989–997. https://doi.org/10.1093/emboj/16.5.989
Blanchetot C, Jonge ND, Desmyter A, Ongenae N, Hofman E, Klarenbeek A, Sadi A, Hultberg A, Kretz-Rommel A, Spinelli S, Loris R, Cambillau C, Haard HD (2016) Structural mimicry of receptor interaction by antagonistic interleukin-6 (IL-6) antibodies. J Biol Chem 291:13846–13854. https://doi.org/10.1074/jbc.M115.695528
Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L (1998) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J 334:297–314. https://doi.org/10.1042/bj3340297
Paonessa G, Graziani R, Serio AD, Savino R, Ciapponi L, Lahm A, Salvati AL, Toniatti C, Ciliberto G (1995) Two distinct and independent sites on IL-6 trigger gp 130 dimer formation and signalling. Embo J 14:1942–1951. https://doi.org/10.1002/j.1460-2075.1995.tb07186.x
Boulanger MJ, Chow DC, Brevnova EE, Garcia KC (2003) Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science 300:2101–2104. https://doi.org/10.1126/science.1083901
Chalaris A, Schmidt-Arras D, Yamamoto K, Rose-John S (2012) Interleukin-6 trans-signaling and colonic cancer associated with inflammatory bowel disease. Dig Dis 30:492–499. https://doi.org/10.1159/000341698
Garbers C, Heink S, Korn T, Rose-John S (2018) Interleukin-6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discov 17:395–412. https://doi.org/10.1038/nrd.2018.45
Atal S, Fatima Z (2020) IL-6 Inhibitors in the treatment of serious COVID-19: a promising therapy? Pharmaceut Med 34:223–231. https://doi.org/10.1007/s40290-020-00342-z
Kishimoto T (2021) IL-6: from arthritis to CAR-T-cell therapy and COVID-19. Int Immunol 33:515–519. https://doi.org/10.1093/intimm/dxab011
Copaescu A, Smibert O, Gibson A, Phillips EJ, Trubiano JA (2020) The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J Allergy Clin Immunol 146:P518-534. https://doi.org/10.1016/j.jaci.2020.07.001
Mai TT, Nguyen PG, Le MT, Tran TD, Huynh PNH, Trinh DT, Nguyen QT, Thai KM (2022) Discovery of small molecular inhibitors for interleukin-33/ST2 protein-protein interaction: a virtual screening, molecular dynamics simulations and binding free energy calculations. Mol Divers 26:2659–2678. https://doi.org/10.1007/s11030-021-10359-4
Meng XY, Zhang HX, Mezei M, Cui M (2012) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157. https://doi.org/10.2174/157340911795677602
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074-1082. https://doi.org/10.1093/nar/gkx1037
Molecular Operating Environment (MOE) (2015) Version 2015.10. Chemical Computing Group Inc., Montreal
Kalai M, Montero-Julian FA, Grötzinger J, Fontaine V, Vandenbussche P, Deschuyteneer R, Wollmer A, Brailly H, Content J (1997) Analysis of the human interleukin-6/human interleukin-6 receptor binding interface at the amino acid level: proposed mechanism of interaction. Blood 89:1319–1333
The Protein Data Bank. https://www.rcsb.org/structure/4ZS7. Accessed 20 June 2021
LeadIT. Version 2.1.8. https://www.biosolveit.de/LeadIT. Accessed 15 Sep 2021
Ngo TD, Tran TD, Le MT, Thai KM (2016) Computational predictive models for P-glycoprotein inhibition of in-house chalcone derivatives and drug-bank compounds. Mol Divers 20:945–961. https://doi.org/10.1007/s11030-016-9688-5
Sekhar PN (2021) Computational prediction of hERG blockers using homology modelling, molecular docking and QuaSAR studies. Results Chem 3:100101. https://doi.org/10.1016/j.rechem.2021.100101
Delaney JS (2004) ESOL: estimating aqueous solubility directly from molecular structure. J Chem Inf Comput Sci 44:1000–1005. https://doi.org/10.1021/ci034243x
Daina A, Zoete V (2016) A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11:1117–1121. https://doi.org/10.1002/cmdc.201600182
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Tran TS, Le MT, Tran TD, Tran TH, Thai KM (2020) Design of curcumin and flavonoid derivatives with acetylcholinesterase and beta-secretase inhibitory activities using in silico approaches. Molecules 25:3644. https://doi.org/10.3390/molecules25163644
Lindahl E, Abraham MJ, Hess B, Spoel Dvd (2021) GROMACS 2021.02 Documentation (Release 2021.02). Zenodo. https://doi.org/10.5281/zenodo.4457591
Mackerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Matto C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616. https://doi.org/10.1021/jp973084f
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368. https://doi.org/10.1002/jcc.21816
Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4:17. https://doi.org/10.1186/1758-2946-4-17
Bepari AK, Reza HM (2021) Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation. PeerJ 9:e11261. https://doi.org/10.7717/peerj.11261
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Nguyen PT, Yu H, Keller PA (2014) Discovery of in silico hits targeting the nsP3 macro domain of chikungunya virus. J Mol Model 20:2216. https://doi.org/10.1007/s00894-014-2216-6
Kumari R, Kumar R, Lynn A (2014) g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH, Hou T (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 119:9478–9508. https://doi.org/10.1021/acs.chemrev.9b00055
Homeyer N, Gohlke H (2012) Free energy calculations by the molecular mechanics Poisson-Boltzmann surface area method. Mol Inform 31:114–122. https://doi.org/10.1002/minf.201100135
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10:449–461. https://doi.org/10.1517/17460441.2015.1032936
Chéron N, Shakhnovich EI (2017) Effect of sampling on BACE-1 ligands binding free energy predictions via MM-PBSA calculations. J Comput Chem 38:1941–1951. https://doi.org/10.1002/jcc.24839
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Ghose AK, Viswanadhan VN, Wendoloski JJ (1999) A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem 1:55–68. https://doi.org/10.1021/cc9800071
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. https://doi.org/10.1021/jm020017n
Egan WJ, Merz KM, Baldwin JJ (2000) Prediction of drug absorption using multivariate statistics. J Med Chem 43:3867–3877. https://doi.org/10.1021/jm000292e
Muegge I, Heald SL, Brittelli D (2001) Simple selection criteria for drug-like chemical matter. J Med Chem 44:1841–1846. https://doi.org/10.1021/jm015507e
Ali SA, Hassan MI, Islam A, Ahmad F (2014) A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr Protein Pept Sci 15:456–476. https://doi.org/10.2174/1389203715666140327114232
Lobanov MY, Bogatyreva NS, Galzitskaya OV (2008) Radius of gyration as an indicator of protein structure compactness. Mol Biol 42:623–628. https://doi.org/10.1134/S0026893308040195
Oyewusi HA, Huyop F, Wahab RA (2022) Molecular docking and molecular dynamics simulation of Bacillus thuringiensis dehalogenase against haloacids, haloacetates and chlorpyrifos. J Biomol Struct Dyn 40:1979–1994. https://doi.org/10.1080/07391102.2020.1835727
Lauria A, Tutone M, Ippolito M, Pantano L, Almerico AM (2010) Molecular modeling approaches in the discovery of new drugs for anti-cancer therapy: the investigation of p53-MDM2 interaction and its inhibition by small molecules. Curr Med Chem 17:3142–3154. https://doi.org/10.2174/092986710792232021
Tran TT, Tran QH, Nguyen QT, Le MT, Trinh DT, Thai KM (2022) Identification of potential interleukin-8 inhibitors acting on the interactive site between chemokine and CXCR2 receptor: a computational approach. PLoS ONE 17:e0264385. https://doi.org/10.1371/journal.pone.0264385
Tran QH, Nguyen QT, Vo NQ, Mai TT, Tran TT, Tran TD, Le MT, Trinh DT, Thai KM (2022) Structure-based 3D-Pharmacophore modeling to discover novel interleukin 6 inhibitors: an in silico screening, molecular dynamics simulations and binding free energy calculations. PLoS ONE 17:e0266632. https://doi.org/10.1371/journal.pone.0266632
Thai KM, Ngo TD, Tran TD, Le MT (2013) Pharmacophore Modeling for Antitargets. Curr Top Med Chem 13:1002–1014. https://doi.org/10.2174/1568026611313090004
Mak KK, Pichika MR (2019) Artificial intelligence in drug development: present status and future prospects. Drug Discov Today 24:773–780. https://doi.org/10.1016/j.drudis.2018.11.014
Acknowledgements
This work was supported by the VietNam National Foundation for Science and Technology Development (NAFOSTED) under the Grant Number 108.05-2018.15 (to Quoc-Thai Nguyen).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tran, QH., Nguyen, QT., Tran, TT.N. et al. Identification of small molecules as potential inhibitors of interleukin 6: a multi-computational investigation. Mol Divers 27, 2315–2330 (2023). https://doi.org/10.1007/s11030-022-10558-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10558-7