Abstract
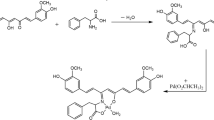
A new ligand FIPB = 5-(1H-imidazo[4,5-f][1,10]phenanthrolin-2-yl)furan-2-yl-2-boronic acid, having three cobalt(III) polypyridyl complexes [Co(phen)2(FIPB)]3+(1) {FIPB = 5-(1H-imidazo[4,5-f][1,10]phenanthrolin-2-yl)furan-2-yl-2-boronic acid}, (phen = 1,10-Phenanthroline), [Co(bpy)2(FIPB)]3+(2) (bpy = 2,2’bipyridyl), [Co(dmb)2(FIPB)]3+(3) (dmb = 4, 4′-dimethyl 2, 2′-bipyridine) have been synthesized and characterized by elemental analysis, ES-MS,1H-NMR, 13C-NMR, UV-Vis and FTIR. Their DNA binding behavior has been explored by various spectroscopic titrations and viscosity measurements, which indicated that all the complexes bind to calf thymus DNA by means of intercalation with different binding strengths. The binding properties of these all three complexes towards calf-thymus DNA (CT-DNA) have been investigated by UV-visible, emission spectroscopy and viscosity measurements.The experimental results suggested that three Co(III) complexes can intercalate into DNA base pairs,but with different binding affinities. Photo induced DNA cleavage studies have been performed and results indicate that three complexes efficiently cleave the pBR322-DNA in different forms. The three synthesized compounds were tested for antimicrobial activity by using Staphylococcus aureus and Bacillus subtilis organisms, these results indicated that complex 1 was more activity compared to other two complexes against both tested microbial strains. The in vitro cytotoxicity of these complexes was evaluatedby MTT assay, and complex 1 shows higher cytotoxicity than complex 2 and 3 on HeLa cells.









Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.All data generated or analyzed during this study are included in this published article.
References
S.P. Fricker, (2007) Metal based drugs: from serendipity to design. Dalton Trans, 4903-4917
Knoll JD, Turro C (2015) Control and utilization of ruthenium and rhodium metal complex excited states for photoactivated cancer therapy. Coord Chem Rev 282-283:110–126
Leung CH, Zhong HJ, Chan DSH, Ma DL (2013) Bioactive iridium and rhodium complexes as therapeutic agents. Coord Chem Rev 257:1764–1776
Gitlin JD, Lill R (2012) Special issue: cell biology of metals. BiochimicaetBiophysicaActa 1823:1405
Howerton BS, Heidary DK, Glazer EC (2012) Strained ruthenium complexes are potent light-activated anticancer agents. J Am Chem Soc 134(20):8324–8327
Frezza M, Hindo S, Chen D, Davenport A, Schmitt S, Tomco D, Dou QP (2010) Novel metals and metal complexes as platforms for cancer therapy. Curr Pharm Des 16:1813–1825
Matos CS, de Carvalho ALMB, Lopes RP, Marques MPM (2012) New strategies against prostate cancer–Pt (II)-based chemotherapy. Curr Med Chem 19:4678–4687
Mokaberi P, Babayan-Mashhadi F, Tehranizadeh ZA, Saberi MR, Chamani J (2020) Analysis of the interaction behaviour between Nano-Curcumin and two human serum proteins: combining spectroscopy and molecular stimulation to understand protein-protein interaction. J Biomol Struct Dyn:1–20. https://doi.org/10.1080/07391102.2020.1766570
Sharifi-Rad A, Mehrzad J, Darroudi M, Saberi MR, Chamani J (2020) Oil-in-water nanoemulsions comprising Berberine in olive oil: biological activities, binding mechanisms to human serum albumin or holotransferrin and QMMD simulations. J Biomol Struct Dyn 39:1029–1043. https://doi.org/10.1080/07391102.2020.1724568
Shakibapour N, Sani FD, Beigoli S, Sadeghian H, Chamani J (2018) Multi-spectroscopic and molecular modeling studies to reveal the interaction between propyl acridone and calf thymus DNA in the presence of histone H1: binary and ternary approaches. J Biomol Struct Dyn 37:359–371. https://doi.org/10.1080/07391102.2018.1427629
Kamshad M, Talab MJS, Beigoli S, Rad AS, Chamani J (2018) Use of spectroscopic and zeta potential techniques to study the interaction between lysozyme and curcumin in the presence of silver nanoparticles at different sizes. J Biomol Struct Dyn 37:2030–2040. https://doi.org/10.1080/07391102.2018.1475258
Mokaberi P, Reyhani V, Amiri-Tehranizadeh Z, Saberi MR, Beigoli S, Samandara F, Chaman J (2019) New insights into the binding behavior of lomefloxacin and human hemoglobin using biophysical techniques: binary and ternary approaches. New J Chem 43:8132. https://doi.org/10.1039/c9nj01048c
Pyle AM, Long EC, Barton JK (1989) Shape-selective targeting of DNA by phenanthrenequinonediiminerhodium(III) photocleaving agents. J Am Chem Soc 111(12):4520–4522
Sitlani A, Long EC, Pyle AM, Barton JK (1992) DNA photocleavage by phenanthrenequinonediimine complexes of rhodium(III): shape-selective recognition and reaction. J Am Chem Soc 114(7):2303–2312
Jin L, Yang P (1997) Synthesis and DNA binding studies of CoIII mixed-ligand complex containing dipyrido [3,2-a: 2′,3′-c]phenazine and phen. Polyhedron 16:3395–3398
Song Y-M, Lu X, Yang M, Zheng X-R (2005) Study on the interaction of platinum(IV), gold(III) and silver(I) ions with DNA, transition met. Chem. 30:499–502
Chai L-Q, Huang J-J, Zhang H-S, Zhang Y-L, Zhang J-Y, Li Y-X (2014) An unexpected cobalt(III) complex containing a Schiff base ligand: synthesis, crystal structure, spectroscopic behavior, electrochemical property and SOD-like activity. SpectrochimActa A 131:526–533
Thamilarasan V, Sengottuvelan N, Sudha A, Srinivasan P, Chakkaravarthi G (2016) Cobalt(III) complexes as potential anticancer agents: physicochemical, structural, cytotoxic activity and DNA/protein interactions. J Photochem Photobiol B 162:558–569
Kumar LS, Prasad KS, Revanasiddappa HD (2012) Synthesis, spectroscopic characterization, antimicrobial, DNA binding and oxidative-induced DNA cleavage activities: New oxovanadium (IV) complexes of 2-(2-hydroxybenzylideneamino) isoindoline-1, 3-dione. SpectrochimActa A 97:659–666
Rodrigues ARO (2014) Benzothienoquinolines: new one-pot synthesis and fluorescence studies of their interaction with DNA and polynucleotides. J Photochem Photobiol A 294:20–30
Nagababu P, Satyanarayana S (2007) DNA binding and cleavage properties of certain ethylenediamine cobalt (III) complexes of modified 1, 10-phenanthrolines. Polyhedron 26:1686–1692
Tanzadehpanah H, Asoodeh A, Saberi MR, JamshidkhanChamani (2013) Identification of a novel angiotensin-I converting enzyme inhibitory peptide from ostrich egg white and studying its interactions with the enzyme. Innov Food Sci Emerg Technol 18:212–219
Moosavi-Movahedi AA, Golchin AR, Nazari K, Chamania J, Saboury AA, Bathaie SZ, Tangestani-Nejad S (2004) Microcalorimetry, energetics and binding studies of DNA–dimethyltin dichloride complexes. Thermochimica Acta 414:233–241
Vuradi RK, Putta VR, Nancherla D, Sirasani S (2016) Luminescent behavior of Ru(II) Polypyridyl morpholine complexes, synthesis, characterization, DNA, protein binding, sensor effect of ions/solvents and docking studies. J Fluoresc 26:689–701
Sani FD, NiloufarShakibapour SB, Sadeghian H, Hosainzadeh M, Chamani J (2018) Changes in binding affinity between ofloxacinand calf thymus DNA in the presence of histone H1: spectroscopic and molecular modeling investigations. J Lumin 203:599–608. https://doi.org/10.1016/j.jlumin.2018.06.083
NavaneethaNambagari SP, Vuradi RK, Satyanarayana S (2019) Study of the interaction of co(III) Polypyridyl complexes with DNA: an experimental and computational approach. Nucleosides Nucleotides Nucleic Acids 38(6):400–417
Ravi C, Vuradi RK, Avudoddi S, Ramchander M, Satyanarayana S (2019) Induction of apoptosis in SKOV3 cells and DNA binding by Cobalt(III) polypyridyl complex. Russ J Bioorgchem+ 45(4):273–284
Vuradi RK, NavaneethaNambigari PP, Gopu S, Kotha LR, Deepika G, Vinoda Rani M, SatyanarayanaSirasani (2020) Study of anti-apoptotic mechanism of ruthenium(II) polypyridyl complexes via RT-PCR and DNA binding. ApplOrganometal Chem 34:5332. https://doi.org/10.1002/aoc.5332
Perrin D, Annarego WLF, Perrin DR (1980) Purification of laboratory chemicals, second edn. Pergamon Press, New York
Marmur JA (1961) A procedure for the isolation of deoxyribonucleic acid from micro organisms. J MolBiol 3:208–218
Yamada M, Tanaka Y, Yoshimoto Y, Kuroda S, Shimo I (1992) Synthesis and properties of diamino-substituted dipyrido [3,2-a:2′,3′-c] phenaxcopyzine. Bull Chem Soc Jpn 65:1006–1011
Vlcek AA (1967) Preparation of Co(dipy)2X2+ complexes (X- = chloride, bromide, iodide, nitrite) by controlled oxidative processes. Inorg Chem 6(7):1425–1427
Steck EA, Day AR (1943) Reactions of phenanthraquinone and retenequinone with aldehydes and ammonium acetate in acetic acid solution. J Am ChemSoc 65:452–456
Wolfe A, Shimer GH, Mechan T (1987) Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 26:6392–6396
Mc Ghee JD, Von Hippel PH (1974) Theoretical aspects of DNAprotein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J MolBiol 86:469–489
Rashidipour S, Naeeminejad S, Chamani J (2015) Study of the interaction between DNP and DIDS with human hemoglobin as binary and ternary systems: spectroscopic and molecular modeling investigation. J Biomol Struct Dyn 34:57–77. https://doi.org/10.1080/07391102.2015.1009946
Brown KN, Geue RJ, Hambley TW, Hockless DCR, Rae AD, Sargeson AM (2003) Specificity in template syntheses of hexaaza-macrobicyclic cages: [Pt(Me5-tricosatrieneN6)]4+ and [Pt(Me5-tricosaneN6)]4+. Org Biomol Chem 1:1598–1608
Bernhardt, P. V., Bramley, R., Geue, R. J., Ralph, S. F., Sargeson, A. M., (2007) An expanded cavity hexaamine cage for copper(ii), Dalton Trans., 1244–1249
Comba P, Martin B, Sanyal A (2013) An efficient fluctuating charge model for transition metal complexes. J Comput Chem 34:1598–1608
Bol JE, Buning C, Comba P, Reedijk J, Strohle M (1998) Molecular mechanics modeling of organic backbone of metal-free and coordinated ligands. J Comput Chem 19:512–523
Hypercube Hyper chem. 7.5. Alberta, Canada, (2002)
Rajender Reddy M, Venkat Reddy P, Praveen Kumar Y, Srishailam A, Navaneetha N, Satyanarayana S (2014) Synthasis, characterization, DNA binding, light switch "on and off", cytotoxicity, sensor and docking studies of ruthenium(II) and cobalt(III) Polypyridyl complexes. J Fluoresc 24:803–817
Navaneetha N, Ramasree D, Kiran Kumar M, Uma V, Nagababu P, Satyanarayana S (2013) Molecular dynamic simulations of Co(III) and Ru(II) Polypyridyl complexes and docking studies with dsDNA. Medicinal Chemistry Research 22(11):5557–5565
Grippo AL, Lucidi S (1997) A globally convergent version of the Polak-Ribière conjugate gradient method. Math Program 78(3):375–391
Venugopal R, Mariappan M, Eringathodi S, Mallayan P, Vaiyapuri SP, Mohammad AA (2008) Non-covalent DNA binding and cytotoxicity of certain mixed-ligand ruthenium(ii) complexes of 2,2′-dipyridylamine and diimines. Dalton Trans:2157–2170
Mariappan M, Bhaskar MG (2005) Effects of Anthracene and Pyrene units on the interactions of novel Polypyridylruthenium(II) mixed-ligand complexes with DNA. Eur J Inorg Chem 2164-2173
Satyanarayana S, Dabrowiak JC, Chaires JB (1992) Neithernor Λ-Tris(phenanthroline)ruthenium(II) binds to DNA by classical intercalation. Biochemistry 31:9319–9324
Nagababu P, Shilpa M, Latha JNL, Ira B, Srinivas PNBS, Paveen Kumar Y, Reddy KL, Satyanarayana S (2011) Synthesis, characterization, DNA binding properties, fluorescence studies and toxic activity of cobalt(III) and ruthenium(II) polypyridyl complexes. J Fluoresc 21:563–572
Barton JK, Raphael AL (1984) Photoactivated stereospecific cleavage of double-helical DNA by cobalt(III) complexes. J Am Chem Soc 106:2466–2468
Hay BP (1993) Methods for molecular mechanics modeling of coordination compounds. CoordChemRev 126(1–2):177–236
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:363–367
Acknowledgements
The authors are grateful to CFRD, Osmania University forspectral analysis.
Funding
The authors are grateful to the UGC-UPE(FAR) ProgramOsmania University, Hyderabad, for funding.
Author information
Authors and Affiliations
Contributions
The author SPconceived this research and designed experiments, wrote the paper and participated in the revisions of it; RKV participated in the design and interpretation of the data helped in writing the paper and participated in the revision of it; SG and VR are participated in analysis of data and revision of paper; NN investigated the molecular docking and interpretation of data; SSN Supervision the research, designed experiments correction of manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
This article does not contain any studies involving human participants performed by any of the authors and does not contain any studies involving animals performed by any of the authors.
Conflicts of Interest
“The authors declare that they have no potential conflict of interest in relation to the study in this paper”.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Perka, S., Vuradi, R.K., Gopu, S. et al. Influence of Co(III) Polypyridyl Complexes on Luminescence Behavior, DNA Binding, Photocleavage, Antimicrobial Activity and Molecular Docking Studies. J Fluoresc 31, 1009–1021 (2021). https://doi.org/10.1007/s10895-021-02727-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-021-02727-y