Abstract
Nuclear tandemly repeated ribosomal RNA genes (18S-5.8S-25S rDNA and 5S rDNA) have been proven to be excellent cytogenetic markers for karyotype analysis of various higher plants by using the fluorescence in situ hybridization (FISH) technique. To illustrate physical mapping of these rDNAs in brown seaweed, Saccharina japonica, chromosomes, an approximately 5400-bp transcription unit of 18S-5.8S-25S rDNA was assembled by cloning the 25S rDNA after paired-end sequencing of two screened clones from a bacterial artificial chromosome library of kelp gametophytes. In contrast to the conserved 18S-5.8S-25S rDNA and ITS1 in S. japonica, the 245-bp ITS2 was variable in sequence. The cloned 5S rDNA revealed that the 120-bp conserved coding region was separated by a diverse intergenic spacer sequence that were 250, 445, 905, or 1335 bp in length. On average, the 18S-5.8S-25S rDNA of kelp female and male gametophytes had 45 and 41 copies per haploid genome, respectively, as detected by quantitative real-time PCR, whereas the 5S rDNA had 2590 and 2648 copies, respectively. Southern hybridization with labeled probes of 18S rDNA or 5S rDNA demonstrated that kelp gametophytes possessed only one locus each of the 18S-5.8S-25S or 5S rDNAs. This was further confirmed by FISH analysis using the same labeled probes, thus illustrating that 18S-5.8S-25S rDNA is located at the intercalary region of chromosome 23, whereas 5S rDNA at the sub-telomeric region of chromosomes 27. The localization of these rDNAs using the FISH technique has facilitated the identification of individual chromosomes and karyotype analysis of this kelp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The previous cytogenetic reports on the economically significant brown seaweed, Saccharina japonica (Aresch.) C. E. Lane, C. Mayes, Druehl et G. W. Saunders (=Laminaria japonica Aresch.), have demonstrated that kelp chromosomes are not only very small (0.57–2.61 μm) in size but also nearly homomorphic in shape (Yabu 1973; Tai and Fang 1976, 1977; Yabu and Yasui 1991; Zhou et al. 2004; Liu et al. 2012a,b). These chromosomal features thus make it difficult to identify individual chromosomes and analyze the karyotype of brown seaweeds (Lewis 1996) by use of conventional cytogenetic techniques. However, using the sensitive fluorescence in situ hybridization (FISH) technique, especially with nuclear ribosomal RNA (rRNA) genes as probes, has been proven to be efficient for the investigation of species with small or similarly sized chromosomes (Leitch and Heslop-Harison, 1992; Garcia et al. 2012). Thus, this enables the identification and discrimination of different chromosomes within and between chromosome complements (Hamon et al. 2009).
The nuclear rRNA genes, which occur in nearly all of eukaryotes in the form of tandemly repeated arrays (Long and Dawid 1980; Rogers and Bendich 1987; Maluszynska et al. 1998; Volkov et al. 2007; Torres-Machorro et al. 2010; Garcia et al. 2012), are categorized into two different families. The major rRNA family, which constitutes the nucleolus organizer regions (NORs), contains the 18S, 5.8S, and 25S/26S/28S rRNA genes that encode the 45S pre-ribosomal RNA as synthesized by RNA polymerase I (Mandal 1984; Sollner-Webb and Tower 1986; Hemleben and Zentgraf 1994; Paule and White 2000). The minor non-nucleolus forming 5S rRNA family, which is transcribed by RNA polymerase III (Mandal 1984; Sollner-Webb and Tower 1986; Paule and White 2000), contains a highly conserved 120-bp long coding region and a variable intergenic spacer (IGS) region (Long and Dawid 1980; Drouin and Moniz de Sá 1995; Barciszewska et al. 2000; Volkov et al. 2007). Due to their universal presence in eukaryotic genomes, the number, position, and structure of the 5S and 18S-5.8S-25S rDNA loci are considered important characteristics of a given species, genus, or group (Garcia et al. 2012). As we know, the sequences of 18S rDNA (Bi et al. 2009), internal transcribed spacer (ITS) 1 and ITS2 (Yotsukura et al. 1999), and 5S rDNA (Yotsukura et al. 2002) in S. japonica and other closely related species have been extensively characterized. However, the molecular organization of these two families of rDNAs, copy number, and physical mapping on chromosomes yet remain to be completed.
To characterize the rRNA genes in S. japonica, these were cloned by polymerase chain reaction (PCR), followed by the assembly of the gene structure based on their gene sequences. The copy number of rRNA genes in the haploid gametophytes of S. japonica was estimated by use of quantitative real-time PCR (Q-RT PCR), as previously employed in bacteria (Zeng and Jiao 2009), copepods (Zagoskin et al. 2010), fungi (Black et al. 2013), tea plants (Xu et al. 2015), and dinoflagellates (Saito et al. 2002; Galluzzi et al. 2004). Finally, the precise location of the 18S-5.8S-25S and 5S rRNA genes was determined by using the FISH technique. To our knowledge, this is the first report on the physical mapping of rDNAs in the kelp S. japonica or even in algae except for the dinoflagellates, Alexandrium spp. (Figueroa et al. 2014). These data will be useful for the construction of physical maps of chromosomes, the integration of genetic and physical maps, the discrimination of whole chromosomes for karyotypic analysis, the identification of U/V sex chromosomes (Bachtrog et al. 2011), and the comparison of chromosomal organization in phylogenetic studies on S. japonica and other related species in the future.
Materials and methods
Algal strain and culture
The Rongfu strain of Saccharina japonica was selected as the algal material in the present study. Female and male gametophyte clones germinated from zoospores were isolated according to cell size (about 20 μm for females and 10 μm for males) under a microscope and cultured in 500-mL flasks under vegetative growth conditions of 30 μmol photons m−2 s−1 at 17 ± 1 °C, with a photoperiod of 12 h: 12 h light: dark as previously described (Zhou and Wu 1998). During culture, the flasks were shaken several times by hand at intervals each day, and the Provasoli’s enriched seawater (PES) medium (Starr and Zeikus 1993) was replaced every 2 weeks.
DNA extraction
Genomic DNA was extracted from freshly harvested gametophytes according to the modified cetyltrimethyl ammonium bromide (CTAB) method of Hu and Zhou (2001). The extracted DNA was dissolved in Tris-EDTA (TE) buffer which was composed of 10 mM Tris-HCl and 1 mM EDTA at pH = 8.0.
Cloning of the 18S-5.8S-25S and 5S rRNA genes
Amplification of the 18S rRNA gene using the previously designed primers 18S-F/18S-R (Bi et al. 2009) (Table 1) was conducted in a 25-μL reaction volume containing the template DNA (50 ng), Mg2+ (2.5 mM), Taq DNA polymerase (1 U), dNTPs (100 μM each), forward and reverse primers 18S-F/18S-R (0.2 μM each), and 1× reaction buffer. PCR was performed in a gradient Mastercycler (Eppendorf, Germany) and programmed as described by Hu and Zhou (2001) except for the use of 30 cycles of the reaction and annealing at 58 °C for 45 s. The amplified products were resolved on a 1.0% low-melting-point agarose gel for DNA recovery. The target product was purified using a UNIQ-10 column DNA gel extraction kit (Sangon, China) and ligated into a pMD19-T vector (TaKaRa, China). The constructed vector was subsequently transformed into Escherichia coli DH5α competent cells (Leihao, Shanghai, China), and several positive clones were screened and sent to Sangon (Shanghai, China) for sequencing. Because ITS sequences are generally less conserved (Coleman 2003; Calonje et al. 2009), more than 20 clones were sequenced following S. japonica ITS1 and ITS2 cloning.
According to the above-verified sequence of the 18S rRNA gene, one pair of primers 18S-F1/18S-R1 (Table 1) was designed by using the Primer Premier 5.0 software (http://www.premierbiosoft.com/primerdesign/index.html) and employed in PCR screening the constructed bacterial artificial chromosome (BAC) library of the kelp female gametophyte for the 18S-5.8S-25S rRNA genes. The PCR reaction system and program was the same as described earlier except for the template of the kelp genomic DNA, which was replaced by BAC clone DNA. Two clones (BAC Clones L10 and E18), which possibly contained the 18S-5.8S-25S rRNA genes, were screened. Based on the partial sequence of the 25S rRNA gene after paired-end sequencing of these two clones, one pair of primers 25S-F/25S-R (Table 1) was designed for cloning of the 25S rRNA gene. Furthermore, two pairs of primers, 18S-F2/ITS2-R and ITS1-F/25S-R1 (Table 1), were designed and used in cloning the ITS1-5.8S-ITS2 rRNA genes of the kelp haploid gametophytes. The PCR reaction system and program was also the same as that for the 18S rRNA gene, except for the extension time used for amplification with the pairs of primers 18S-F1/18S-R1, 25S-F/25S-R, 18S-F2/ITS2-R, and ITS1-F/25S-R1 which was 1, 5, 4, and 4 min, respectively.
The genomic DNA of the 5S rRNA gene was amplified with the reported pair of primers, 5Ss-F/5Ss-R (Table 1) (Yotsukura et al. 2002). Each 25-μL PCR reaction system was composed of template DNA (50 ng), Mg2+ (2.5 mM), Taq DNA polymerase (1 U), dNTPs (100 μM), forward and reverse primers 5Ss-F/5Ss-R (0.2 μM each), and 1× reaction buffer. PCR amplification was programmed as described by Hu and Zhou (2001), with minor modifications, in which 30 cycles and annealing at 62 °C for 45 s were employed. Another pair of primers, 5S-F/5S-R (Table 1), was used to amplify the IGS of the S. japonica 5S rRNA gene. The PCR reaction system and program was the same as that for cloning the 5S rRNA gene, except for the use of a 2-min extension time. The amplified products were resolved on a 1.0% low-melting-point agarose gel for DNA recovery, cloning, and sequencing as earlier described.
Bioinformatics analysis
The DNAMAN (Lynnon Biosoft, USA) and BioEdit (Hall 1999) softwares were employed for sequence assembly, alignment, and restriction site analysis. BLASTP analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to obtain the homologous sequences of the 18S, 5.8S, and 25S rRNA genes from Costaria costata, Saccharina angustata, Ectocarpus siliculosus, and Laminaria digitata. The ITS and 5S rDNA sequences of several brown seaweeds were retrieved from NCBI. Supplementary Table 1 presents the details of these genes.
Copy number estimation of the rRNA genes using Q-RT PCR
Ribosomal RNA gene copies in S. japonica haploid gametophytes were estimated by using Q-RT PCR as previously described (Zagoskin et al. 2010; Xu et al. 2015). Prior to Q-RT PCR amplification, two vectors, pMD19-T18S and pMD19-T5S, which contained 1716-bp- and 120-bp-long 18S and 5S rDNAs, respectively, were constructed as references as described above. Both plasmid DNAs were extracted and digested by HindIII into linear plasmids for the construction of a calibration curve. This curve was plotted based on a series of 10-fold dilutions of digested pMD19-T18S (from approximately 30 ng to 3 pg) or pMD19-T5S (from 2.75 × 109 to 2.75 × 105 copies) DNA against the resulting fluorescence intensity (threshold cycle, CT) as detected by Q-RT PCR. The copy number per fg plasmid DNA molecule (N p) was calculated using the equation:
N p = N A × m/M (Zagoskin et al. 2010);
where N A is the Avogadro constant, m is the DNA amount (i.e., 1 fg) for which the copy number is calculated, and M is the molar mass of the plasmid. Because 1 bp of DNA corresponds to 660 g mol−1, the N p of the 18S rRNA gene is approximately 207 and that of the 5S rRNA gene is 315. Each plasmid DNA dilution was conducted in triplicate. The concentration of the initial plasmid DNA or the total S. japonica gametophyte DNA was determined by using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA).
The primers used in the Q-RT PCR for the 18S-5.8S-25S rRNA (q18SF/q18SR) and 5S rRNA (q5SF/q5SR, q5ST) genes are also shown in Table 1, and the amplification of the 18S-5.8S-25S rRNA gene was performed on an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, USA) using the SYBR RT-PCR kit (TaKaRa) according to the manufacturer’s recommendations. After pre-denaturation at 95 °C for 15 s, the amplification was programmed for 40 cycles of 94 °C for 5 s, 60 °C for 20 s, and a ramp of 0.5 °C/30 s to 95 °C. The Q-RT PCR reactions for 5S rDNA comprised the template DNA (200 ng), Mg2+ (50 μM), Taq DNA polymerase (5 U μL−1), dNTPs (5 μM each), forward and reverse primers q5SF/q5SR (10 μM each), TaqMan probe q5ST (5 μM), and RNase-free H2O to 20 μL. TaqMan probe q5ST was used because we could not design a specific pair of primers such as q5SF/q5SR (Table 1) for the 5S rRNA gene due to its too short insert (120 bp only) in the plasmid pMD19-T5S. This reaction program was as follows: 1 cycle of pre-denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. All Q-RT PCR reactions were performed in triplicate.
The copy number of rDNA in the kelp haploid genome was calculated using the following equation:
N r = N m × N p × C/k (Zagoskin et al. 2010);
where N r is the 18S rDNA or 5S rDNA copy number, C is the size of the S. japonica haploid genome (C = 0.5975 pg) as estimated by Ye et al. (2015), N m is the experimentally determined initial amount of the 18S or 5S rRNA gene templates, N p is the same as earlier described, and k is the amount of genomic DNA template of the sample that was used in the Q-RT PCR.
Southern blot analysis
To obtain more information on the 18S-5.8S-25S and 5S rDNAs of the kelp gametophytes, Southern blot analysis was conducted as described by Ye et al. (2014). In this experiment, we chose 18S rDNA (synthesized by using the primer pair, 18S-F/18S-R, Table 1) and 5S rDNA (synthesized by using the primer pair, 5Ss-F/5Ss-R, Table 1) as probes. Aliquots of isolated DNA (approximately 2 μg per sample) were digested to completion at 37 °C for 12–14 h independently by using NcoI and SalI. Agarose electrophoresis of the digested DNA samples, blotting onto a positively charged nylon membrane (Pall, USA), hybridization with the synthesized 18S and 5S rDNA probes, and signal visualization on an XBT-1 film (Kodak, USA) were performed as previously described (Ye et al. 2014).
FISH analysis
Chromosomes were prepared from S. japonica gametophytes according to previous studies (Liu et al. 2012a,b). Fresh female and male gametophytes of S. japonica were separately treated with 0.02% colchicine for 8–10 h at room temperature. After incubation, the samples were washed and then fixed in freshly prepared Carnoy’s fixative solution (100% ethanol/acetic acid, 3:1, v/v) for 24 h (Schweizer 1976). Chromosome squashes were subsequently prepared following the enzymatic softening of material as described elsewhere (Liu et al. 2012b). After screening under a phase contrast microscope, the well-spread mitotic chromosomes were pre-treated sequentially with 100 mg mL−1 of RNase A for 1 h and 0.25 mg mL−1 pepsin for 15 min (Lim et al. 2001), subjected to FISH detection, and then counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
The FISH experiment was conducted using two DNA probes, namely, 18S and 5S rDNAs, which were 1824-bp and 120-bp fragments labeled with digoxigenin (DIG)-12-dUTP (Roche, Switzerland) after PCR amplification from S. japonica. Hybridization was performed as described by Lim et al. (2001) with slight modifications. The slides were UV cross linked for 2 min, washed in 2 × SSC (3 × 5 min), and rinsed in a series of 70, 95, and 100% ethanol for 2 min each time and air dried. Approximately 10 μL of the hybridization mixture, which contained 50% formamide (v/v), 2 × SSC (0.3 M NaCl, 0.03 M sodium citrate, pH 7), 5% dextran sulfate (w/v), and 5 ng μL−1 of the probe, was added to the slide, covered with a slip of plastic sheet, heated to 100 °C in a water bath for 5 min, and then transferred to a 37 °C air oven for hybridization for 12 h. Post-hybridization washes of the slides were performed twice in a slightly more stringent solution of 2 × SSC at 42 °C in a shaker for 10 min to remove unbound, nonspecifically bound, or weakly hybridized probe. To detect the hybridization sites, the slides were washed with 4 × SSC and then blocked with 30 μL of 5% bovine serum (w/v) in PBS per slide, covered with plastic coverslips, and then incubated at 37 °C for 30 min. The two probes were detected by incubating with anti-digoxigenin-rhodamine conjugated to digoxigenin in a blocking reagent (Roche, Switzerland) at 37 °C for 1 h. After incubation, the slides were washed with 4 × SSC twice, each for 5 min (Osuji et al. 2006), and then counterstained with DAPI (2 μg mL−1). Fluorescence images were captured using a Leica DM4000 epifluorescent microscope with an Orca ER camera and then processed using Adobe Photoshop 7.0 (http://www.adobe.com/products/photoshop.html) by uniformly adjusting the contrast and brightness (Liu et al. 2012a).
Results
Characterization of the 18S-5.8S-25S and 5S rRNA genes in S. japonica
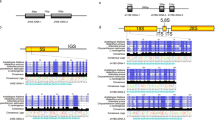
Based on partial sequencing of two screened clones from the constructed BAC library of the female S. japonica gametophytes, several pairs of primers (Table 1) were designed to clone the coding sequence of 18S-5.8S-25S rRNA gene cluster via PCR amplification (Supplementary Fig. 1). One transcription unit (Fig. 1) of this gene cluster in S. japonica was assembled using the sequences of the amplified products. Approximately 5400 bp of the coding sequence of 18S-5.8S-25S rDNAs was confirmed by using other re-designed primers (data not shown) different from the used ones for gene cloning and was then deposited to NCBI (GenBank Accession No. KX827269). The length of the 18S rRNA gene was 1824 bp, and its sequence was identical to the reported 18S rDNA of the same species (GenBank Accession No. EU293553, Bi et al. 2009) and highly similar to those that were respectively cloned from Laminaria digitata, Saccharina angustata, Costaria costata, and Ectocarpus siliculosus (Supplementary Table 1).
The 5.8S rRNA gene of S. japonica was 160 bp in size and it was identical in size to those of C. costata (Bhattcharya and Druehl 1988) and E. siliculosus (Peters et al. 2010). Only two bases differed from that of S. angustata, L. digitata, C. costata, and E. siliculosus, all of which also belong to the same phylum Phaeophyta, thereby indicating the conservation of the 5.8S rDNA in various kelp species (Supplementary Table 1).
The 25S rRNA gene of S. japonica was 2891 bp in length, and sequence alignment with four other brown algal species (C. costata, S. angustata, E. siliculosus, and L. digitata, Supplementary Table 1) showed that the nucleotide arrangement of this gene was also highly conserved. The lowest similarity was 97%, which was observed between S. japonica and E. siliculosus.
Sequencing of ITS amplicons revealed that the ITS1 region was 238 bp in length and was identical to that of the other kelp species S. augustata, S. religiosa, and S. ochotensis, with only two different bases in S. diabolia and S. longissma (Supplementary Table 1), which was indicative of sequence conservation. Compared to ITS1, the cloned ITS2 of S. japonica was less conserved in sequence, with a 98% sequence similarity to each clone, although these were of the same length (245 bp).
The 5S rDNA of S. japonica was cloned by PCR amplification using previously reported primers (Table 1, Yotsukura et al. 2002). The size of the PCR products that were amplified by using the primer pair 5S-F2R2 ranged from 300 to 1500 bp (Supplementary Fig. 2). The four distinct bands were subsequently cloned and sequenced. Database searching result showed that each repetitive unit of 5S rDNA included a 120-bp coding region, which was similar to that reported by Yotsukura et al. (2002) but separated by a more diverse IGS (Fig. 1) as determined by sequencing. This variable IGS of S. japonica 5S rDNA was not only in sequence but also in length. Besides the reported length of about 250 bp (Yotsukura et al. 2002), this study has determined that the length of IGSs was 445, 905, and 1335 bp (Supplementary Fig. 2). This finding was confirmed by searching against the recently released kelp genome database by Ye et al. (2015). The IGSs detected in this study were deposited to NCBI (GenBank Accession Nos. KY440189–KY440192), and were characterized by a conserved TATA box that was located −27 to −25 bp upstream of the 120-bp coding sequence as reviewed by Volkov et al. (2004). The 5′ end of the IGSs also contained a block of T-rich sequence, thereby suggesting that this region could be involved in transcription termination as proposed by Korn (1982). Accordingly, these 445-bp, 905-bp, and 1335-bp 5S rDNA IGSs might be potentially used as molecular markers for discriminating between the Chinese and Japanese kelp populations.
Copy number estimation of 18S-5.8S-25S and 5S rRNA gene repeats
Upon the establishment of a standard Q-RT PCR protocol after optimization using various primers and PCR reagent concentrations, a standard curve (Supplementary Fig. 3) with the linearized pMD19-T18S DNA as a template was generated. Linear regression analysis revealed a positive relationship between the C T values and the serially diluted plasmid DNA levels (R 2 = 0.999) (Supplementary Fig. 3). The C T value of the S. japonica 18S rRNA gene in the haploid genome was also detected by Q-RT PCR (Table 2). By referring to the constructed standard curve and the recently reported genome size of S. japonica (Ye et al. 2015), the copy number of 18S-5.8S-25S rRNA gene in S. japonica female and male gametophytes was evaluated (Table 2). The copy number ranged from 39 to 51 (mean ± SD = 45 ± 7, n = 9) and from 22 to 56 (mean ± SD = 41 ± 15, n = 9) per haploid genome of female and male gametophytes, respectively (Table 2). Although the female and male gametophytes showed fluctuations in 18S-5.8S-25S rRNA gene copy number, these were not statistically different from each other (Student’s t-test, P = 0.701 > 0.05).
Similar to the estimation for the 18S rRNA gene but using a designed TaqMan probe (q5ST, Table 1) and primers q5SF/q5SR (Table 1), a protocol for the quantification of the 5S rRNA gene copy number was established. A standard control curve between the C T values and the series of 10-fold dilutions of copy number of pMD19-T5S (from 2.75 × 109 to 2.75 × 105 copies) with an R 2 = 0.999 was also constructed (Supplementary Fig. 4). By using this calibration curve, the 5S rRNA gene copy number in S. japonica gametophytes was calculated, which ranged from 1333 to 4404 (mean ± SD = 2590 ± 1399, n = 9) and from 1968 to 3314 (mean ± SD = 2648 ± 595, n = 9) per haploid genome of male and female gametophytes, respectively (Table 2). The female and male gametophytes showed no significant differences (Student’s t-test, P = 0.957 > 0.05) in terms of copy number.
Locus and physical mapping of the 18S-5.8S-25S and 5S rRNA genes
To further investigate the locus of the nuclear rDNAs, genomic DNA that was extracted from S. japonica gametophytes was digested with two different restriction enzymes (SalI and NcoI) and subjected to Southern blot analysis using labeled probes specific to the 1716-bp 18S or 212-bp 5S rRNA genes. Only one hybridization signal was observed in Fig. 2, implying that there might be one locus of these two genes in the S. japonica gametophyte genome that was digested by SalI. When the restriction enzyme NcoI was used to digest the haploid genome of S. japonica, the same result was obtained for the 5S rRNA gene (Fig. 2, lower panel), whereas two signals (Fig. 2, upper panel) were observed for the 18S-5.8S-25S rRNA gene cluster. The intense band, which was about 3.5 kb in size (Fig. 2, upper panel), coincided with our predicted, which was cleaved by NcoI at both sites of 343 and 3980 bp downstream of the coding sequence of the 18S rRNA gene. The size of the minor signal was about 0.8 kb (Fig. 2, upper panel), which was suggestive of an NcoI cleavage site in the IGS sequence upstream of the 18S rRNA gene. Because the IGS sequence of the 18S-5.8S-25S rRNA genes was not cloned in this study, this cleavage site was not confirmed. Nevertheless, the results of Southern blotting analysis (Fig. 2) illustrated that only one locus of both the 18S-5.8S-25S and 5S rRNA genes was present in the haploid genome of the S. japonica gametophytes. Furthermore, no difference in rRNA gene locus between the kelp female and male gametophytes was observed by using Southern blot.
Agarose electrophoresis profile (left) of SalI-digested and NcoI-digested genomic DNA of the Saccharina japonica gametophytes and its corresponding Southern blot pattern (right) hybridized with the biotin-labeled 18S rDNA (upper panel) or 5S rDNA (lower panel). M Marker IV DNA standard, DNA undigested Saccharina japonica genomic DNA, NcoI genomic DNA digested by NcoI, SalI genomic DNA digested by SalI
To determine the 18S-5.8S-25S and 5S rDNA loci on kelp chromosomes in both number and position, at least 100 metaphase nuclei of the S. japonica gametophytes were examined by using the FISH technique. The chromosome number of the haploid gametophytes was 31 (Fig. 3), which was in agreement with the recently reported results of Liu et al. (2012a,b). Rhodamine hybridization signals on the chromosomes counterstained by DAPI are shown in Fig. 3. Of the 20 examined metaphase nuclei that were examined to determine the locus of the 18S-5.8S-25S rRNA genes, 12 nuclei showed only one distinguishable fluorescent signal (Fig. 3, upper panel). On the other hand, 20 of the 30 examined nuclei showed one distinguishable 5S rRNA gene fluorescent signal (Fig. 3, lower panel). These FISH images cytogenetically demonstrated that both 18S-5.8S-25S and 5S rRNA genes were located at one locus on the kelp chromosomes, and these genes were thus encoded by the kelp nuclear genome as well. No difference in locus was observed between the female and male gametophytes by using the FISH technique.
The arrangement of the kelp chromosomes according to decreasing size (Fig. 4) showed that the 18S-5.8S-25S rDNA was located at the intercalary region of chromosome 23, as indicated by the hybridization of the labeled 18S rDNA probe, whereas the 5S rRNA gene was located at the sub-telomeric region of chromosomes 27. The localization of the 18S-5.8S-25S and 5S rDNAs on different chromosomes supported the hypothesis that the latter was not linked to the former in S. japonica (Yotsukura et al. 2002).
FISH images of Saccharina japonica chromosomes in Fig. 3 arranged according to the decreasing length, showing the physical location of the 18S (upper) and the 5S (lower) rDNA sites
Discussion
In addition to the reported sequences of 18S rDNA (Bi et al. 2009), ITS1 and ITS2 (Yotsukura et al. 1999), and 5S rDNA (Yotsukura et al. 2002), this study cloned the coding sequence of the 25S rDNA so as to complete the rDNA repeat unit in the haploid gametophytes of S. japonica. The 18S-5.8S-25S rRNA gene cluster is about 5400 bp long (Fig. 1), and all the sequences of the 18S rDNA, ITS1, 5.8S, and 25S rDNA are highly conserved, whereas that of ITS2 varied. The ITS2 heterogeneity showed both inter-specific variations as suggested by Yotsukura et al. (1999), as well as intra-specific (within S. japonica) variations. Our findings also support the results of Coleman (2003) that there are variations among different ITS2 repeats that exist in a single eukaryote genome. This variation may be mainly due to nucleotide indels and substitutions in parents and then is inherited only in hybrids or polyploids as proposed by Buckler et al. (1997), Volkov et al. (2007), and Poczai and Hyvönen (2010). These findings thus reflect the possible hybrid of Rongfu kelp strain used in this study, which is consistent with the fact that this strain was cross-bred intra-specifically using a female gametophyte clone from Fujian (South of China) and a male gametophyte clone from the Strain Yuanza 10 (cultivated in Shandong, North of China) (Liao et al. 2010). The geographic distance of these two above-mentioned locations from north to south is about 1200 km, indicating the possibility of factors, such as temperature, to affect the genetic variations of its parents so that this hybrid could exhibit obvious heterosis (Liao et al. 2010). Therefore, our findings also imply that implications of ITS2 sequences in evolutionary studies should be performed with caution (Álvarez and Wendel 2003; Poczai and Hyvönen 2010; Wang et al. 2015).
The results of the present study support the findings of Rogers and Bendich (1987), Poczai and Hyvönen 2010, and Torres-Machorro et al. (2010) that the 18S-5.8S-25S rRNA gene cluster is separated by an IGS (Fig. 1). Although the kelp IGS sequence of this major rDNA could not be cloned in the present study, its length could be inferred from the Southern blot profiles (Fig. 2). In the upper panel of Fig. 2, the SalI-digested genomic DNA hybridized with a labeled probe of 1716-bp 18S rDNA shows that the size of a repeat unit of the 18S-5.8S-25S rRNA gene cluster would be within the range of 7.0–8.0 kb. After the removal of the assembled 5400-bp coding sequence of the 18S-5.8S-25S rRNA gene cluster, the remaining 1.6–2.6 kb may highly likely be the IGS sequence. However, further studies that would validate our inferences are warranted.
In contrast to the unknown IGS of the major rDNA, sequencing of the IGS of 5S rDNA was completed in this study, thus unveiling the length and sequence heterogeneity of the kelp 5S rDNA (Supplementary Fig. 2). Interestingly, this particular heterogeneity was not observed in the Japanese populations of S. japonica (Yotsukura et al. 2002). Within a single genome, different size classes of 5S rDNA repeat units were detected in other organisms such as Triticum spp. (Baum and Appels 1992), Populus deltoides (Negi et al. 2002), and Beta vulgaris subsp. maritima (Turner and Brown 2005), but not in kelps. Volkov et al. (2007) suggested that different lengths of the 5S rDNA repeat units in the same genome possibly originated from duplications and rearrangements. Our current understanding of the mechanism of the 5S rDNA IGS inheritance is limited while compared to that of the 18S-5.8S-25S rDNA in plants, because the latter plays an important role in nucleolus construction (Volkov et al. 2007; Poczai and Hyvönen 2010). Based on the size of the cloned 5S rDNA IGS of S. japonica, we infer that the 445-bp IGS was generated from the 250-bp IGS by duplication, and the resulting 445-bp IGS had given rise to the 906-bp and 1335-bp IGSs using the same genomic mechanism. Consequently, we propose that this heterogeneity in the Chinese kelp population probably resulted from duplications and rearrangements of the Japanese kelp 250-bp IGS during acclimatization to the local sea environment in China, possibly since its colonization in the 1920s. However, further investigations in this proposal are needed.
To meet the enormous demand for rRNA in cells, the rDNA gene family occurs as tandem repeats in most eukaryotes (Long and Dawid 1980; Rogers and Bendich 1987; Torres-Machorro et al. 2010). Higher plants usually have from 250 to 20,000 copies per haploid cell (Rogers and Bendich 1987), whereas several species of algae carry from 3 to 2000 copies per haploid cell (Table 3). By using the Q-RT PCR technique, a valid approach for dinoflagellate rRNA copy number estimation as described by both Saito et al. (2002) and Galluzzi et al. (2004), we determined that the 18S-5.8S-25S rDNA consists of 43 repeats, whereas the 5S comprises 2618 repeats (Table 2) on average in the haploid gametophytes of S. japonica regardless of their gender. This copy number is nearly within the range of available documented data on algae (Table 3). Nevertheless, it is necessary to point out that a huge variation occurs in each experiment (Table 2), probably resulting in a huge standard deviation, because different growth phases and environments have been reported to affect the copy number of rRNA genes in a plant (for review, Long and Dawid 1980). This might be the most plausible reason that in the same species, for example, Euglena gracilis, the reported copy number of rRNA genes differs in different documents (Table 3).
Prokopowich et al. (2003) provided the first convincing evidence of a strong positive relationship between genome size and rDNA copy number by surveying 94 species of animals and 68 species of plants. In contrast to this finding, the copy number of rRNA genes in S. japonica seems not very appropriate for its genome size of about 545 Mb (Ye et al. 2015), although the copy number still stands within the range of the available data from algae (Table 3). In these studies on the estimation of yeast rRNA gene copy number, Ide et al. (2010) and Kobayashi (2011) found that the loss of these repeats in yeast cells confers sensitivity to DNA damage caused by mutagens such as UV radiation. Taking this into consideration and the fact that S. japonica can survive under medium irradiance of no more than 30% of the water surface solar irradiance (Fei et al. 1989), which is generally accepted as the result of natural selection and adaptation to low light in the sub-littoral region (Häder and Figueroa 1997), the low copies of rDNA serves as a possible explanation why this particular kelp species is unable to tolerate high irradiance. Evidently, additional experimental evidence is necessary to further support this relationship.
In the present study, Southern blot (Fig. 2) and FISH (Fig. 3) analyses have indicated the presence of one single locus for the 18S-5.8S-25S rRNA genes is detectable in the haploid gametophytes of S. japonica. In the majority of organisms, the 18S-5.8S-25S rRNA genes are on chromosomes that associate with the nucleolus (Griffor et al. 1991; Volkov et al. 2007). Because there is only one prominent nucleolus in the kelp nucleus (Bouck 1965; Dai and Fang 1979), this locus is supposed to be confined to the nucleolus organizer region (NOR). Although the sites of the 18S-5.8S-25S rRNA genes showed a preferential distribution on the short arms of chromosomes (Roa and Guerra 2012), we were unable to determine whether this locus was located on the short arm of chromosome 23 due to the indistinguishable position of centromere (Figs. 3 and 4).
Unlike the nuclear 18S-5.8S-25S rRNA genes, the 5S rRNA gene is encoded also by plant chloroplast or mitochondrial genomes (Barciszewska et al. 2000; Valach et al. 2014). To avoid this possibility, the cloned 5S rRNA gene sequence from S. japonica was searched against the kelp chloroplast (GenBank Accession No. JQ405663, Wang et al. 2013) and mitochondrion (GenBank Accession No. AP011493, Yotsukura et al. 2010) genomic data on NCBI. No sequences homologous to our cloned 5S rRNA gene were identified, thereby suggesting that the 5S rRNA gene of S. japonica is encoded by the nuclear genome. This has been verified by the FISH image (Fig. 3, lower panel) of the kelp 5S rRNA gene.
The first report on the linkage between the 5S rRNA and 18S-5.8S-25S rRNA genes in the brown seaweed Scytosiphon lomentaria (Kawai et al. 1995) has sparked subsequent studies on other closely related species, and most of the studied Chromophyta, including brown seaweeds, have been shown to harbor a 5S-linked type of rDNA organization (Kawai et al. 1997). Nevertheless, PCR analysis has indicated that the 5S rRNA gene could be distinguished from the 18S-5.8S-25S rDNA in S. japonica (Yotsukura et al. 2002), and the FISH technique employed in this present study has shown that the 5S rRNA gene is located on chromosome 27, which is different from that of the 18S-5.8S-25S rRNA gene (Fig. 4). These findings are in agreement with the results of most plants (Garcia et al. 2012), wherein the use of dual-color FISH for co-localization of the 5S and 18S-5.8S-25S rDNAs on kelp chromosomes will illustrate the unlinked relationship between these two loci.
References
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29:417–434
Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N (2011) Are all sex chromosomes created equal? Trends Genet 27:350–357
Barciszewska MZ, Szymański M, Erdmann VA, Barciszewski J (2000) 5S ribosomal RNA. Biomacromolecules 1:297–302
Baum BR, Appels R (1992) Evolutionary change at the 5S DNA loci of species in the Triticeae. Plant Syst Evol 183:195–208
Bhattcharya D, Druehl LD (1988) Phylogenetic comparison of the small-subunit ribosomal DNA sequence of Costaria costata (Phaeophyta) with those of other algae, vascular plants and oomycetes. J Phycol 24:539–543
Bi Y-H, Zou D-Y, Zhou Z-G (2009) Cloning and sequence analysis of 18S rRNA gene from Laminaria japonica gametophytes and its application as an internal standard. Acta Agriculturae Boreali-Sinica 24:49–54
Black J, Dean T, Byfield G, Foarde K, Menetrez M (2013) Determining fungi rRNA copy number by PCR. J Biomol Tech 24:32–38
Bouck GB (1965) Fine structure and organelle associations in brown algae. J Cell Biol 26:523–537
Buckler ES IV, Ippolito A, Holtsford TP (1997) The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics 145:821–832
Calonje M, Martín-Bravo S, Dobeš C, Gong W, Jordon-Thaden I, Kiefer C, Kiefer M, Paule J, Schmickl R, Koch MA (2009) Non-coding nuclear DNA markers in phylogenetic reconstruction. Plant Syst Evol 282:257–280
Coleman AW (2003) ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet 19:370–375
Curtis SE, Rawson JRY (1981) Characterization of the nuclear ribosomal DNA of Euglena gracilis. Gene 15:237–247
Dai JX, Fang ZX (1979) Cell division of the female gametophytes and juvenile sporophytes of Laminaria japonica. Periodical of Ocean University of China 9:123–127
Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piégu B, Ball SG, Ral J-P, Bouget F-Y, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A 103:11647–11652
Drouin G, Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol 12:481–493
Fei X, Jiang B, Ding M, Wu Y, Huang R, Li H (1989) Light demands of juvenile Laminaria japonica. Chin J Oceanol Limnol 7:1–9
Figueroa RI, Bravo EGI (2010) The use of flow cytometry for species identification and life-cycle studies in dinoflagellates. Deep-Sea Res II 57:301–307
Figueroa RI, Cuadrado A, Stüken A, Rodríguez F, Fraga S (2014) Ribosomal DNA organization patterns within the dinoflagellate genus Alexandrium as revealed by FISH: life cycle and evolutionary implications. Protist 165:343–363
Galluzzi L, Penna A, Bertozzini E, Vila M, Garcés E, Magnani M (2004) Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl Environ Microbiol 70:1199–1206
Garcia S, Garnatje T, Kovařík A (2012) Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121:389–394
Griffor MC, Vodkin LO, Singh RJ, Hymowitz T (1991) Fluorescent in situ hybridization to soybean metaphase chromosomes. Plant Mol Biol 17:101–109
Gruol DJ, Haselkorn R (1976) Counting the genes for stable RNA in the nucleus and chloroplasts of Euglena. Biochim Biophys Acta 447:82–95
Häder DP, Figueroa FL (1997) Photoecophysiology of marine macroalgae. Photochem Photobiol 66:1–14
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamon P, Siljak-Yakovlev S, Srisuwan S, Robin O, Poncet V, Hamon S, de Kochko A (2009) Physical mapping of rDNA and heterochromatin in chromosomes of 16 Coffea species: a revised view of species differentiation. Chromosom Res 17:291–304
Hemleben V, Zentgraf U (1994) Structural organization and regulation of transcription by RNA polymerase I of plant nuclear ribosomal RNA genes. In: Nover L (ed) Plant promoters and transcription. Springer, Berlin, pp 3–24
Howell SH (1972) The differential synthesis and degradation of ribosomal DNA during the vegetative cell cycle in Chlamydomonas reinhardtii. Nature 240:264–267
Hu Y-J, Zhou Z-G (2001) Extraction of RAPD-friendly DNA from Laminaria japonica (Phaeophyta) after enzymatic dissociation of the frozen sporophyte tissue. J Appl Phycol 13:415–422
Ide S, Miyazaki T, Maki H, Kobayashi T (2010) Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327:693–696
Kawai H, Muto H, Fujii T, Kato A (1995) A linked 5S rRNA gene in Scytosiphon lomentaria (Scytosiphonales, Phaeophyceae). J Phycol 31:306–311
Kawai H, Nakayama T, Inouye I, Kato A (1997) Linkage of 5S ribosomal DNA to other rDNAs in the chromophytic algae and related taxa. J Phycol 33:505–511
Keller M, Tessier LH, Chan RL, Weil JH, Imbault P (1992) In Euglena, spliced-leader RNA (SL-RNA) and 5S rRNA genes are tandemly repeated. Nucleic Acids Res 20:1711–1715
Kobayashi T (2011) Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci 68:1395–1403
Korn LJ (1982) Transcription of Xenopus 5S ribosomal RNA genes. Nature 295:101–105
Leitch IJ, Heslop-Harison JS (1992) Physical mapping of the 18S-5.8S-26S rDNA genes in barley by in situ hybridization. Genome 35:1013–1018
Lewis RJ (1996) Chromosomes of the brown algae. Phycologia 35:19–40
Liao L, Lai X, Chen W, Luo D, Li C, Du H (2010) Study on a new species of kelp (Rongfu kelp) farming southward test in Shantou. Fish Sci Technol Z1:12–14
Lim KB, Wenneckes J, de Jong JH, Jacobsen E, van Tuyl JM (2001) Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridisation. Genome 44:911–918
Liu Y, Bi Y-H, Gu J-G, Li L-H, Zhou Z-G (2012a) Localization of a female-specific marker on the chromosomes of the brown seaweed Saccharina japonica using fluorescence in situ hybridization. PLoS One 7:e48784
Liu Y, Bi Y-H, Zhou Z-G (2012b) Karyological observation on Saccharina japonica chromosomes stained with DAPI. J Fish China 36:50–54
Long EO, Dawid IB (1980) Repeated genes in eukaryotes. Annu Rev Biochem 49:727–764
Maluszynska J, Hasterok R, Weiss H (1998) rRNA genes-their distribution and activity in plants. In: Maluszynska J (ed) Plant cytogenetics. Wydawnictwo Uniwersytetu Śląskiego, Katowice, Poland, pp 75–95
Mandal RK (1984) The organization and transcription of eukaryotic ribosomal RNA genes. Prog Nucleic Acid Res Mol Biol 31:115–160
Maruyama S, Misumi O, Ishii Y, Asakawa S, Shimizu A, Sasaki T, Matsuzaki M, Shin-i T, Nozaki H, Kohara Y, Shimizu N, Kuroiwa T (2004) The minimal eukaryotic ribosomal DNA units in the primitive red alga Cyanidioschyzon merolae. DNA Res 11:83–91
Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–251
Negi MS, Rajagopal J, Chauhan N, Cronn R, Lakshmikumaran M (2002) Length and sequence heterogeneity in 5S rDNA of Populus deltoids. Genome 45:1181–1188
Osuji JO, Okoli BE, Heslop-Harrison JSP (2006) Cytology and molecular cytogenetics of Cucumeropsis mannii Naudin: implications for breeding and germplasm characterization. Int J Bot 2:187–192
Parrow MW, Burkholder JM (2002) Flow cytometric determination of zoospore DNA content and population DNA distribution in cultured Pfiesteria spp. (Pyrrhophyta). J Exp Mar Biol Ecol 267:35–51
Paule MR, White RJ (2000) Transcription by RNA polymerases I and III. Nucleic Acids Res 28:1283–1298
Peters AF, van Wijk SJ, Cho GY, Scornet D, Hanyuda T, Kawai H, Schroeder DC, Cock JM, Boo SM (2010) Reinstatement of Ectocarpus crouaniorum Thuret in Le Jolis as a third common species of Ectocarpus (Ectocarpales, Phaeophyceae) in Western Europe, and its phenology at Roscoff, Brittany. Phycol Res 58:157–170
Poczai P, Hyvönen J (2010) Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol Biol Rep 37:1897–1912
Prokopowich CD, Gregory TR, Crease TJ (2003) The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48–50
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225
Rogers SO, Bendich AJ (1987) Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Saito K, Drgon T, Robledo JA, Krupatkina DN, Vasta GR (2002) Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl Environ Microbiol 68:5394–5407
Scott NS (1973) Ribosomal RNA cistrons in Euglena gracilis. J Mol Biol 83:327–336
Siu C-H, Chiang K-S, Swift H (1976) Characterization of cytoplasmic and nuclear genomes in the colorless alga Polytoma III. Ribosomal RNA cistrons of the nucleus and leucoplast. J Cell Biol 69:383–392
Sollner-Webb B, Tower J (1986) Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem 55:801–830
Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58:307–324
Spring H, Grierson D, Hemleben V, Stöhr M, Krohne G, Stadler J, Franke WW (1978) DNA contents and numbers of nucleoli and pre-rRNA-genes in nuclei of gametes and vegetative cells of Acetabularia mediterranea. Exp Cell Res 114:203–215
Starr RC, Zeikus JA (1993) UTEX-the culture collection of algae at the University of Texas at Austin 1993 list of cultures. J Phycol 29(Suppl. 2):1–106
Tai CH, Fang TC (1976) Some preliminary observations on the parthenogenesis in Laminaria japonica Aresch. Acta Genet Sinica 3:32–38
Tai CH, Fang TC (1977) The chromosomes of Laminaria japonica Aresch. Acta Genet Sinica 4:325–328
Torres-Machorro AL, Hernández R, Cevallos AM, López-Villaseñor I (2010) Ribosomal RNA genes in eukaryotic microorganisms: witnesses of phylogeny? FEMS Microbiol Rev 34:59–86
Turner DJ, Brown TA (2005) Abridged 5S rDNA units in sea beet (Beta vulgaris subsp. maritima). Genome 48:352–354
Valach M, Burger G, Gray MW, Lang BF (2014) Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res 42:13764–13777
Volkov RA, Medina FJ, Zentgraf U, Hemleben V (2004) Molecular cell biology: organization and molecular evolution of rDNA, nucleolar dominance, and nucleolus structure. Prog Bot 65:106–146
Volkov RA, Komarova NY, Hemleben V (2007) Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst Biodivers 5:261–276
Wang X, Shao Z, Fu W, Yao J, Hu Q, Duan D (2013) Chloroplast genome of one brown seaweed, Saccharina japonica (Laminariales, Phaeophyta): its structural features and phylogenetic analyses with other photosynthetic plastids. Mar Genomics 10:1–9
Wang X-C, Liu C, Huang L, Bengtsson-Palme J, Chen HM, Zhang J-H, Cai DY, Li J-Q (2015) ITS1: a DNA barcode better than ITS2 in eukaryotes? Mol Ecol Resour 15:573–586
Xu J, Xu Y, Yonezawa T, Li L, Hasegawa M, Lu F, Chen J, Zhang W (2015) Polymorphism and evolution of ribosomal DNA in tea (Camellia sinensis, Theaceae). Mol Phylogenet Evol 89:63–72
Yabu H (1973) Alternation of chromosomes in the life history of Laminaria japonica Aresch. Bull. Fac. Fish Hokkaido Univ 23:171–176
Yabu H, Yasui H (1991) Chromosome number in four species of Laminaria (Phaeophyta). Jpn J Phycol 39:185–187
Ye N, Zhang X, Miao M, Fan X, Zheng Y, Xu D, Wang J, Zhou L, Wang D, Gao Y, Wang Y, Shi W, Ji P, Li D, Guan Z, Shao C, Zhuang Z, Gao Z, Qi J, Zhao F (2015) Saccharina genomes provide novel insight into kelp biology. Nat Commun 6:6986
Ye R-X, Yu Z, Shi W-W, Gao H-J, Bi Y-H, Zhou Z-G (2014) Characterization of α-type carbonic anhydrase (CA) gene and subcellular localization of α-CA in the gametophytes of Saccharina japonica. J Appl Phycol 26:881–890
Yotsukura N, Denboh T, Motomura T, Horiguchi T, Coleman AW, Ichimura T (1999) Little divergence in ribosomal DNA internal transcribed spacer −1 and −2 sequences among non-digitate species of Laminaria (Phaeophyceae) from Hokkaido, Japan. Phycol Res 47:71–80
Yotsukura N, Kawai T, Motomura T, Ichimura T (2002) Tandem 5S ribosomal RNA genes and the spacer region sequences of three Japanese Laminaria species. J Appl Phycol 14:233–239
Yotsukura N, Shimizu T, Katayama T, Druehl LD (2010) Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol 22:243–251
Zagoskin MV, Marshak TL, Mukha DV, Grishanin AK (2010) Chromatin diminution process regulates rRNA gene copy number in freshwater copepods. Acta Nat 2:52–57
Zeng Y, Jiao N (2009) A novel method for assessment of 16S rRNA gene copy number in bacterial genomes by pulsed-field gel electrophoresis and PCR amplification. J Rapid Meth Autom Microbiol 17:274–279
Zhou LR, Dai JX, Shen SD (2004) An improved chromosome preparation from male gametophyte of Laminaria japonica (Heterokontophyta). Hydrobiologia 512:141–144
Zhou Z-G, Wu C-Y (1998) Clone culture of Laminaria japonica and induction of its sporophytes. Chin J Biotech 14:109–111
Acknowledgements
This research was supported by the National Natural Science Foundation of China (41376136 and 31201992), the National High Technology Research and Development Program of China (2012AA10A406) and Shanghai Universities Plateau Discipline Project of Marine Sciences.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOC 13306 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, L., Yang, QF., Dong, WS. et al. Characterization and physical mapping of nuclear ribosomal RNA (rRNA) genes in the haploid gametophytes of Saccharina japonica (Phaeophyta). J Appl Phycol 29, 2695–2706 (2017). https://doi.org/10.1007/s10811-017-1206-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-017-1206-3