Abstract
The genes encoding ribosomal RNA (rRNA) are the most abundant genes in the eukaryotic genome. They reside in tandem repetitive clusters, in some cases totaling hundreds of copies. Due to their repetitive structure and highly active transcription, the rRNA gene repeats are some of the most fragile sites in the chromosome. A unique gene amplification system compensates for loss of copies, thus maintaining copy number, albeit with some fluctuations. The unusual nature of rRNA gene repeats affects cellular functions such as senescence. In addition, we recently found that the repeat number determines sensitivity to DNA damage. In this review, I would like to introduce a new aspect of the rRNA gene repeat (called rDNA) as a center of maintenance of genome integrity and discuss its contribution to evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Three unique regions stand out in the eukaryotic genome. One is the telomere. Telomeres are located at the ends of chromosomes; they protect the ends from degradation by nucleases and prevent connections with other chromosomal ends. Another is the centromere, a locus that associates with microtubules and functions to segregate chromosomes in mitosis, and is characterized by a huge repetitive structure. Finally, the ribosomal RNA (rRNA) gene repeats, also known as rDNA (ribosomal DNA), also present a unique type of locus in the eukaryotic genome. The rDNA clusters consist of tandem repetitive genes that encode ribosomal RNAs which form the skeletal framework of the ribosome. The rRNA genes are the most abundant and critical housekeeping genes in the cell, and are highly conserved from bacteria to humans. As a repetitive sequence, the rDNA is one of the most fragile regions in the eukaryotic genome and this property affects cellular functions such as senescence [1].
In this review, I would like to introduce a new aspect of rDNA, as a center of genome maintenance. In addition, I will discuss its role as a driver of evolution that affects mutation rates.
rDNA as the “king of the housekeeping genes”
In the budding yeast Saccharomyces cerevisiae, rRNA is transcribed as 35S rRNA and then processed into three mature rRNAs (18S, 5.8S, and 26S, Fig. 1). Together with a small, 5S rRNA, which is transcribed independently from 35S rRNA, these RNA molecules form the skeletal framework of the ribosome [2]. Ribosome is a protein–RNA complex that translates mRNA to protein and it is abundant in the cell. Ribosomal proteins (RP) account for approximately 50% of the total protein and rRNA represents approximately 80% of the total RNA in a yeast cell [3]. To meet this huge biosynthetic demand, eukaryotic cells contain hundreds of copies of rRNA genes organized into clusters. In human cells, there are five rDNA clusters, located in chromosomes 13, 14, 15, 21 and 22. Each cluster contains ~70 copies, bringing the total number of rDNA gene copies to ~350 (~15 Mbp) per haploid genome [4]. In general, plants have more rDNA copies. For example, a pea (Pisum sativum) has ~4,000 copies [5]. In S. cerevisiae, there is a single cluster, comprising approximately 150 copies of rDNA, located on chromosome XII. This cluster covers about 60% of chromosome XII and about 10% of the whole genome [6]. For these reasons, we can reasonably consider the rDNA as the “king of the housekeeping genes” in terms of function and quantity (Fig. 1).
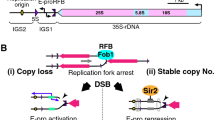
Structure of the budding yeast rDNA locus. The rDNA is a tandem repeating array on chromosome XII. A repeating unit (9.1 kb) has 5S and 35S rRNA genes and two intergenic spacer regions (IGS1, 2). rARS and RFB are the replication origin and replication fork barrier site, respectively. EXP (~500 bp) is an expansion sequence that contains RFB and E-pro. E-pro is a bidirectional promoter for non-coding transcripts that function in the regulation of rDNA repeat numbers. The rDNA structure is broadly conserved from yeast to human, though in the human genome the 5S rDNA is found in independent arrays
rDNA as one of the most fragile sites in the genome
The highly repetitive nature of the rDNA region makes it highly recombinogenic and vulnerable to loss of copies after deleterious recombination events among the repeats. For example, when the repeats sustain damage, it may be repaired by recombination with another copy. In this case, the repeat loses several copies between the damaged site and the template copy for repair (Fig. 2a). In addition, when double-strand break occurs in a repeat, it may be repaired by the single-strand annealing pathway (Fig. 2b, [7]). Unless damaged, the repetitive nature of rDNA leads to the formation of unusual secondary structure due to interaction of DNA strands between repeats. This structure may inhibit DNA replication and the stalled replication fork is repaired through recombination. In this way, the rDNA, due to its repetitive nature, is easily untangled but, in the process, loses copies. Trinucleotide repeat elements are also known as unstable DNA sequences [8]. They make secondary structures that inhibit DNA replication and become “hotspots” of recombination to cause translocation or increase of the repeat number [9]. The repeats usually induce irreversible change in the chromosome. On the contrary, in case of rDNA, by a dedicated rDNA maintenance system, each organism keeps its rDNA copy numbers stable at a characteristic value (Table 1). The main compensatory mechanism utilized by this system is gene amplification (see below, and also reference [10] for review). As a consequence, rDNA copy number varies both downward (loss) and upward (amplification) due to these contractions and expansions. Thus, the rDNA is one of the most dynamic regions in terms of copy number. In other words, it is unstable (or fragile) part of the genome [10].
Repair of damage in the rDNA repeats results in the reduction of copy number. a Recombinational repair between the repeats. During G1, DNA damage in one rDNA repeat may be repaired by recombination with another rDNA repeat. rDNA repeats located between the damaged and template copies may be lost by the recombination. b Single-strand annealing (SSA) pathway for repair of repeating genes. When double-strand break (DSB) occurs in a repeat, single-stranded regions are created adjacent to the break and they extend to the complementary strands. Then the strands anneal to each other to repair. In this case, a copy will be lost
The mechanism to maintain the rDNA copy number
The gene amplification mechanism that counteracts recombination-mediated loss of rDNA copies is well studied in budding yeast [6, 11]. During the S phase of the cell cycle, replication starts from replication origins, and is inhibited at the replication fork barrier site (RFB) by the function of the fork blocking protein, Fob1 (Fig. 3) [12]. This inhibition works as a recombinational hotspot to induce amplification for copy number recovery as follow; The single-stranded region of the blocked structure may be a target for endonuclease activity, leading to the formation of double-strand breaks. The broken end can then be repaired by homologous recombination with a sister chromatid. In the case of a repetitive sequence like the rDNA, the broken end may also be recombined with a neighboring copy unequally, and re-start replication there. This recombinational repair re-replicates several rDNA copies. As a result, the copy number increases in one of the two sister chromatids.
rDNA amplification model. a In normal situations, the silencing protein, Sir2, represses E-pro activity, allowing the cohesin protein complex (dotted ellipse) to associate with the IGS. DSBs are repaired by equal sister chromatid recombination, with no change in rDNA copy number. b In situations where copy number is reduced, Sir2 repression is removed and E-pro is activated. This E-pro transcription displaces cohesin from the IGS. The lack of cohesion means that unequal sister chromatids can be used as templates for repair of DSBs, resulting in changes in rDNA copy number. The gray lines represent single chromatids (double-strand DNA) (see text for the details)
This amplification is regulated by a promoter, E-pro, which directs transcription of a non-protein coding transcript [13]. E-pro is located beside the RFB and starts transcription bi-directionally (Fig. 1) [14, 15]. These transcripts interfere with the function of cohesin, a protein that connects sister chromatids. As a result of the inability of cohesin to link sister chromatids in the presence of E-pro, unequal sister chromatid recombination is increased [13]. When the repeat number is around the wild-type level, E-pro transcription is repressed by Sir2, the NAD+-dependent histone deacetylase. Sir2 alters chromatin structure, and as a result, the amplification does not occur. On the other hand, when rDNA copy number is reduced, Sir2 repression is removed and the amplification is induced [13]. Though it is not known how a cell monitors the rDNA copy number, one possible mechanism is that the auto-regulation of SIR2 gene may be related. If SIR2 itself is regulated by Sir2 protein, when the rDNA copy number reduces, Sir2 proteins that were released from rDNA bind to SIR2 promoter to repress the transcription. As a result, the amount of Sir2 in the cell is reduced and E-pro can start transcription to activate the rDNA amplification system. When the copy number reaches near the wild-type level, Sir2 is spent by the rDNA and the repression of SIR2 gene is reduced. Then the amount of Sir2 increases to repress E-pro and the amplification stops. In fact, in a sir2 mutant, the amplification does not stop, and reaches around 300 copies [11]. Thus, rDNA copy number may be monitored and stably maintained at the proper level (Fig. 3). In addition, not only the number but also the quality of the copies seems to be maintained by the same Fob1-dependent recombination system. It is known that all of the rDNA copies have almost identical sequences [16]. This phenomenon is called “homogenization” [17]. The Fob1-dependent recombination makes it possible to convert (repair) a mutation that occurs in a unit using another unit as an information donor.
Extra-coding functions of rDNA
As the rDNA occupies a large part of the genome, its maintenance indirectly affects cellular functions. I suggest the term “extra-coding functions” for these additional effects of rDNA. Because rDNA is a centrally important element in cellular physiology due to its role in mRNA translation, people have studied its functions from that perspective and such extra-coding functions have not been seriously probed. However, if we consider rDNA as the largest repetitive region in the genome, we are compelled to see its role in a different light.
The maintenance of rDNA consumes numerous factors that are also involved in general genome maintenance, such as replication, recombination, repair, and chromatin modification. Therefore, the rDNA condition (copy number and stability) changes the balance of these factors in the nucleus. One good example is histone modification. The rDNA is known to be a silenced region where non-coding transcripts, such as E-pro, are repressed. This silenced region is equivalent to heterochromatin in higher eukaryotic cells. Sir2, together with Net1 and Cdc14 (called RENT complex) [18, 19], is responsible for the transcriptionally inactive state of the rDNA region. The same histone deacetylase, Sir2, is also responsible for telomere silencing, together with Sir3 and Sir4 [20]. As a result, when the rDNA copy number is reduced, telomere silencing is increased because the silencing factors are released from rDNA and are then free to associate with the telomere [21]. Similarly, other regions where Sir2 associates are also affected by rDNA copy number. Thus, due to the fact that the rDNA occupies a large part of the genome, its copy number affects the effective concentration of various factors that function in genome maintenance.
rDNA instability promotes aging
As mentioned above, reduction of rDNA copy number affects the balance of genomic factors. In addition, the locus is a highly unstable site where the copy number of rDNA is always changing. The instability is also thought to affect cellular functions [1]. One typical example is the effect of rDNA instability on aging. Though budding yeast is a unicellular organism, it exhibits apparent aging phenomena. A “mother” cell produces a smaller daughter cell by a process of asymmetric cell division, called “budding”. There are critical differences between mother and daughter cells, apart from their size. The mother ages as she produces a daughter and she dies after ~20 cell division cycles. In contrast, the daughter rejuvenates and recovers the capability to bud another 20 times. That is, the daughter is equivalent to a progeny (baby) in higher eukaryotic organisms (Fig. 4a).
Asymmetrical cell division in budding yeast. a Life cycle of budding yeast. Budding yeast divides asymmetrically. The mother (bigger) cell ages with each cell division, leading to senescence after ~20 cell cycles. However, the daughter cell (smaller) rejuvenates and maintains the capability for division. b rDNA is unstable in the mother. Defective cellular constituents such as oxidized proteins, vacuoles, episomes, and old mitochondria stay and accumulate in the mother cell. Stable rDNA segregates to the daughter cell while unstable rDNA remains in the mother
The relationship between rDNA and lifespan was first demonstrated by Guarente’s group at MIT [22]. They reported that “pop-out” molecules (named extra-chromosomal rDNA circles, ERCs), derived from the rDNA by recombination, accumulated only in the mother cell, and they proposed that this ERC accumulation induced senescence. They also found that general episomes such as plasmid vectors also accumulated in the mother cell and promote senescence as well. The mechanism is still unknown, but the authors speculated that accumulated ERCs or episomes titrate factors, which are required for maintenance of “youthfulness” in the mother cell. Recently, we established a yeast strain in which replication initiation activity in the rDNA was reduced [23]. In this strain, replication initiation occurs only in the chromosomal rDNA array but not in plasmids, therefore, ERCs did not replicate and, consequently, did not accumulate in the mother cell. We analyzed cellular lifespan in this strain. Contrary to our expectation, these cells exhibited a shortened lifespan. We found that this lifespan shortening depended on rDNA instability, which was only detectable in the mother cell. As rDNA instability is known to increase the numbers of ERCs [24], i.e., it is upstream of ERC accumulation, we postulated that rDNA instability affects lifespan shortening independent of ERC copy number. We also tested the relationships between non-rDNA episomes and lifespan. Interestingly, episomes could also induce rDNA instability. One explanation for this observation is that episomes may titrate factors required for chromosome maintenance (for example, topoisomerase, replication machinery, etc.) and that rDNA stability is sensitive to any shortage of these factors (Fig. 4b).
How rDNA instability leads to cellular senescence is still unknown. One possibility is that unstable rDNA changes the effective concentrations of some proteins, such as repair enzymes. In fact, mutants in DNA repair genes are known to have a shorter lifespan in yeast and human cells [25, 26]. Moreover, aged cells often show genome instability [27]. Therefore, one possible model for the mechanism by which rDNA instability promotes aging is that (1) rDNA instability leads to an accumulation of repair enzymes at the locus because of more recombinational events, (2) The effective free concentration of these factors drops, and as a result, stability of the genome as a whole is reduced, (3) The damage checkpoint control is activated. This reduces cellular functions by blocking the cell cycle, and finally, (4) cells stop growing (Fig. 5, see review [1]).
The rDNA theory for aging. The rDNA is one of the most unstable regions in the genome. Therefore, its instability affects cellular functions. rDNA instability a directly reduces cellular functions through dysfunction of ribosomes, b activates the damage checkpoint control that reduces cellular functions through elongation of cell cycle, c sequesters repair enzymes, resulting in the instability of non-rDNA regions and d the instability of non-rDNA regions reduces cellular functions through the checkpoint control and dysfunction of important genes (see [1])
rDNA copy number determines the sensitivity to DNA damage
Up to this point, I have explained how the rDNA instability affects cellular functions. While the high gene copy numbers observed in eukaryotic rDNA clusters has been interpreted to reflect the demand for large quantities of ribosomes, it is also known, paradoxically, that about half of the rDNA copies are not transcribed in yeast and human cells [28] and in some plants, only a small percentage of the copies is transcribed. This discrepancy between theory and experimental results has been a long-standing puzzle to workers in the field. Recently, we proposed a potential solution to this conundrum [29]. As mentioned above, cells have a unique recombination system for rDNA amplification to keep proper copy number for each species. We found that inhibiting the expression of FOB1 during the amplification allows fixing the rDNA copy number at various levels [6]. In fact, we isolated yeast strains with 20, 40, 60, 80, or 110 (wild-type level) copies of rDNA. In these low-copy-number strains (especially, 20- and 40-copy strains), all of the rDNA repeats were strongly transcribed, and there was no longer any untranscribed copy. We characterized the low-copy-number strains and found that they grew normally, but had increased sensitivity to DNA damage by factors such as ultraviolet radiation and carcinogens. There was a clear negative correlation between copy number and sensitivity to such DNA damaging factors. In other words, the damage sensitivity of the cell was determined by rDNA copy number.
Untranscribed copies are necessary for DNA damage repair
Why does the cell show an increase in sensitivity to DNA damage when the rDNA copy number is reduced? In the low-copy strain, it was found that damaged rDNA is not efficiently repaired in the S phase of the cell cycle. As all of the copies are intensively transcribed in the low-copy strain, we speculated that the transcription is related to the repair defect. In fact, a pol I mutant in which the rDNA is not transcribed did not show any low-copy-number-dependent damage sensitivity. Therefore, rDNA transcription interferes with damage repair. As a direct cause of the repair defect, we identified premature sister chromatid separation in cells with reduced rDNA copies. This premature separation inhibits sister chromatid recombination for damage repair. In fact, establishment of artificial sister-chromatid cohesion was able to prevent the low-copy-number-dependent damage sensitivity. We also found that condensin dissociation led to the premature separation of sister chromatids in low-rDNA-copy-number mutants. In agreement with this observation, Strunnikov and colleagues reported that condensin poorly associated with rDNA in the low-rDNA-copy-number strains [30]. Johzuka and Horiuchi found that pol I transcription hindered condensin association to rDNA [31]. We confirmed these results in our low-copy strains. In accord with these results, condensin is known to be required for damage repair and chromatid cohesion [32, 33].
In summary, due to the lack of untranscribed rDNA in the low-copy strain, there are no available binding sites for condensin. As a result, sister chromatids separate prematurely, before damage repair is completed. In other words, in the wild-type copy number cell, the untranscribed copies are working as a “foothold” where the DNA damage repair enzymes, such as condensin, gather and do their jobs (Fig. 6). This may explain why plants that live under the sun have more rDNA copies than other organisms.
Untranscribed copies repair DNA damage. In the rDNA repeats, about half of the copies are not transcribed. Such untranscribed copies in the rDNA are the “foothold” for condensin, a protein that facilitates DNA repair by mediating sister-chromatid cohesion. The cohesion makes recombinational repair possible (see text for detail)
rDNA instability reduces the overall genome stability
Our studies indicated that the untranscribed copies were necessary for DNA damage repair [29]. In the low-rDNA-copy strains in which all rDNA copies are transcribed, the rDNA becomes increasingly unstable and the cell cannot survive under DNA-damaging conditions. More interestingly, this instability effect is not confined to the rDNA. We found that other chromosomal regions also showed instability in the low-rDNA-copy strains. This may be a consequence of the activities of the repair enzymes (other than condensin) being concentrated on the damaged rDNA (such as IGS regions) to the detriment of other damaged regions for which efficient repair cannot be afforded. This indicates that the rDNA condition affects the balance of enzymes for genome maintenance as I explained above in the section on aging.
Biological roles of the extra-coding functions of rDNA
As explained above, the rDNA has several extra-coding functions. I would like to discuss whether these functions are mere by-products of its high copy number and instability or they have some biological significance.
In terms of the relationship to aging, rDNA stability decreases in the mother cell with each passing cell division cycle, leading to the gradual onset of aging phenomena [23]. Does this rDNA instability–driven senescence have any biological significance? A discussion of the fob1 mutant sheds some light on this question. In the mutant, there is no recombination in the rDNA locus, and as a result, the copy number is stabilized [6]. The mutant grows normally and its lifespan is increased relative to the wild-type, by ~60% [34, 35]. Due to the increase in lifespan, the ratio of aged cells increases and more genome instability is observed in the population [27]. Hence, rDNA-dependent aging has a role to reduce the proportion of abnormal cells in the population. If multicellular organisms have a similar aging mechanism, it may be of even greater importance, where it would be expected to play a role in preventing neoplastic transformations.
As for the low-copy number-dependent damage sensitivity, I speculate it may contribute to adaptation. Because the rDNA is a fragile site, it may easily lose the copies, especially under stress conditions. Due to the low-copy-number-dependent damage sensitivity, the genome becomes unstable and the mutation rate is increased. Such changes may stimulate an increase in genetic variation, and will therefore increase the overall probability of the occurrence of a mutation that increases fitness under any given stress. A beneficial aspect of this increase in genomic instability is that it occurs only when the rDNA copy number is reduced. Once the stress passes, the copy number recovers through amplification, and therefore viability is not permanently affected.
Relationship to evolution
Finally, I would like to think of the evolution of rDNA itself. During evolution, organisms have adapted to changing environments and have increased their diversity. As a result, the number of genes and cell size increased, and the overall complexity of the organism increased along with them. These evolutionary changes are thought to require more ribosomes to support an increased level of protein synthesis. This increase in demand is thought to select for enhanced rDNA transcription. As the functional product of rDNA is RNA, not protein, an amplification step (translation) is missing relative to protein synthesis. One way to increase the amount of product, in the absence of a translation step, is to amplify the DNA template by increasing copy number. It is thought that this type of selective pressure led to the evolution of the rDNA amplification system. Within this hypothesis in which more ribosomes were required for “bigger cells”, rDNA amplification played a central role in facilitating evolutionary processes. Amphibian oogenesis provides a case in point [36]. During oocyte maturation, the cell is enlarged to accommodate maternal factors required in early development. Concomitantly, rDNA is amplified more than 1,000 times. As this amplification occurs extrachromosomally using a rolling circle replication mechanism, the amplified copies are not stably maintained and disappear in the developmental process.
Repetitive genes such as rDNA are well suited to produce large quantities of RNA, but they are hard to repair, as mentioned above. To facilitate repair, in the case of rDNA, cells have evolved extra copies that are not transcribed. These copies could reduce the transcriptional stress and increase efficiency of repair by making a “footing space” for the enzymes as mentioned above. At the same time, however, a higher copy number engenders genome instability. To stabilize such repetitive sequences, cells have developed heterochromatin (silenced chromatin in budding yeast) domains in the rDNA cluster(s). In a lower eukaryote, budding yeast, heterochromatin is not so common except for the rDNA region, suggesting that the yeast rDNA cluster may be a primordial example heterochromatin, which later evolved to encompass other genomic loci. Taken together, the repetitive nature of the rDNA clusters strikes a delicate balance between high production of rRNA and genome instability.
Conclusions
In this review, I have attempted to describe several unique features of rDNA. rDNA wears two hats: one is as a “king” of housekeeping genes, and the other is as a highly repetitive sequence in the genome. The latter feature is troublesome in terms of genome stability. Sometimes the king behaves selfishly because of his absolute power! For example, the rDNA promotes aging through this unstable feature. However, such effects appear to be beneficial for cellular well-being. rDNA-driven aging functions to kill cells before they accumulate lesions that might endanger the organism as a whole. This is extremely important in the prevention of neoplastic transformations in higher eukaryotic cells. Actually, it is known that abnormalities in the nucleolar organizing regions (NORs) are often observed in human cancer cells [37]. In addition, the copy number-dependent damage sensitivity is thought to contribute to environmental adaptability of the population [29, 38].
There are several fundamental outstanding questions. For example, why does rDNA become unstable as the cell divides? How does rDNA instability promote aging? The answers to these questions are critical for our understanding of the relationship between genome stability and senescence.
The study of the extra-coding functions of rDNA is just in its infancy. In the future, I foresee that the answers to these questions could provide insights that may be useful in lowering the incidence of cancers, as well as other aging-related pathologies.
References
Kobayashi T (2008) A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. BioEssays 30:267–272
Watson JD et al (2004) Molecular biology of the gene, 5th edn. CSHL Press, New York
Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24:437–440
Sakai M, Ohta T, Minoshima S, Kudoh J, Wang Y, de Jong PJ et al (1995) Human ribosomal RNA gene cluster: identification of the proximal end containing a novel tandem repeat sequence. Genomics 26:521–526
Ingle J, Timmis JN, Sinclair J (1975) The relationship between satellite deoxyribonucleic acid, ribosomal ribonucleic acid gene redundancy, and genome size in plants. Plant Physiol 55:496–501
Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12:3821–3830
Fishman-Lobell J, Rudin N, Haber JE (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 12:1291–1303
Ashley CT Jr, Warren ST (1995) Trinucleotide repeat expansion and human disease. Annu Rev Genet 29:703–728
Samadashwily GM, Raca G, Mirkin SM (1997) Trinucleotide repeats affect DNA replication in vivo. Nat Genet 17:298–304
Kobayashi T (2006) Strategies to maintain the stability of the ribosomal RNA gene repeats. Genes Genet Syst 81:155–161
Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117:441–453
Kobayashi T (2003) The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23:9178–9188
Kobayashi T, Ganley ARD (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309:1581–1584
Ganley ARD, Hayashi K, Horiuchi T, Kobayashi T (2005) Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc Natl Acad Sci USA 102:11787–11792
Santangelo GM, Tornow J, McLaughlin CS, Moldave K (1988) Properties of promoters cloned randomly from the Saccharomyces cerevisiae genome. Mol Cell Biol 8:4217–4224
Ganley ARD, Kobayashi T (2007) Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191
Dover GA (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–117
Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233–244
Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245–256
Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11:83–93
Michel AH, Kornmann B, Dubrana K, Shore D (2005) Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev 19:1199–1210
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033–1042
Ganley ARD, Ide S, Saka K, Kobayashi T (2009) The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell 35:683–693
Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13:2570–2580
Park PU, Defossez PA, Guarente L (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol Cell Biol 19:3848–3856
Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F et al (1996) Positional cloning of the Werner’s syndrome gene. Science 272:258–262
McMurray MA, Gottschling DE (2003) An age-induced switch to a hyper-recombinational state. Science 301:1908–1911
French SL, Osheim YN, Cioci F, Nomura M, Beyer AL (2003) In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23:1558–1568
Ide S, Miyazaki T, Maki H, Kobayashi T (2010) Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327:693–696
Wang BD, Butylin P, Strunnikov A (2006) Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 5:2260–2267
Johzuka K, Horiuchi T (2007) RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells 12:759–771
Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M (2002) Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197–202
Lam WW, Peterson EA, Yeung M, Lavoie BD (2006) Condensin is required for chromosome arm cohesion during mitosis. Genes Dev 20:2973–2984
Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L (1999) Elimination of replication fork block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3:447–455
Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17:1497–1506
Brown DD, Dawid IB (1968) Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science 160:272–280
Crocker J, Nar P (1987) Nucleolar organizer regions in lymphomas. J Pathol 151:111–118
Paredes S, Maggert KA (2009) Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA 106:17829–17834
Long EO, Dawid IB (1980) Repeated genes in eukaryotes. Annu Rev Biochem 49:727–764
Acknowledgments
I thank Dr. Satoru Ide in our lab for the figure. This work was supported in part by grants-in-aid for Scientific Research (21247003) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kobayashi, T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell. Mol. Life Sci. 68, 1395–1403 (2011). https://doi.org/10.1007/s00018-010-0613-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0613-2