Abstract
High-affinity K+ (HAK)/K+ uptake (KUP)/K+ transporters (KT) play crucial roles in the regulation of cellular K+ levels. However, little is known about these genes in the Rosaceae family. In this study, 56 putative HAK/KUP/KT genes were identified in genome sequences from Pyrus bretschneideri, Fragaria vesca, and Vitis vinifera, 21 of which were from P. bretschneideri (designated PbHAK1-21). HAK/KUP/KTs from these species, as well as from Arabidopsis and rice, were grouped into five major clusters with eight subclades. Whole-genome duplication/segmental duplication and dispersed duplication largely accounts for the expansion of HAK/KUP/KT families in these five species. Orthologous relationships between pear and Arabidopsis genes suggest that some PbHAKs function as high-affinity K+ transporters or mediators of abiotic stress responses. Cis-regulatory motifs upstream of PbHAK genes also suggest that members of this family respond to environment changes. PbHAK2 and PbHAK12 mRNAs are abundant in roots exposed to normal levels of K+ and are rapidly up-regulated under conditions of K+ deficiency, suggesting that they have crucial roles in K+ uptake, especially at low K+ concentrations. PbHAK12 (orthologous to AtHAK5 from Arabidopsis) is predominately localized in the plasma membrane, consistent with a role in mediating K+ uptake. Some PbHAK mRNA levels also change in response to abiotic stresses such as salt, cold, and drought. Our data reveals potential candidate genes for further functional characterization, and may be useful for breeding pear rootstocks that utilize potassium more efficiently.






Similar content being viewed by others
References
Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134:1135–1145
Airoldi CA, Davies B (2012) Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genom 39:157–165
Altunoglu YC, Baloglu P, Yer EN, Pekol S, Baloglu MC (2016) Identification and expression analysis of LEA, gene family members in cucumber genome. Plant Growth Regul 80:225–241
Al-Younis I, Wong A, Gehring C (2015) The Arabidopsis thaliana K+ uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett 589:3848–3852
Ashley MK, Grant M, Grabov A (2006) Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot 57:425–436
Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8:509
Atkinson NJ, Lilley CJ, Urwin PE (2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol 162:2028–2041
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130:784–795
Chang YH, Li H, Cong Y, Lin J, Sheng BL (2012) Characterization and expression of a phytochelatin synthase gene in birch-leaf pear (Pyrus betulaefolia Bunge). Plant Mol Biol Rep 30:1329–1337
Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF, Sze H (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front Plant Sci 3:25
Chen G, Hu Q, Luo L, Yang T, Zhang S, Hu Y, Yu L, Xu G (2015) Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ 38:2747–2765
Chou KC, Shen HB (2010) Plant-mploc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5:e11335
Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry AA, Fizames C, Sentenac H (2010) Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell Mol Life Sci 67:2511–2532
Davies C, Shin R, Liu W, Thomas MR, Schachtman DP (2006) Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J Exp Bot 57:3209–3216
Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14:119–131
Epstein W, Kim BS (1971) Potassium transport loci in Escherichia coli K-12. J Bacteriol 108:639–644
Fu HH, Luan S (1998) AtKUP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10:63–73
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Gierth M, Mäser P (2007) Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581:2348–2356
Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137:1105–1114
Gomez-Porras JL, Benito B, Riaño-Pachón DM, Haro R, Sklodowski K, Rodrígueznavarro A, Ingo Dreyer I (2012) Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front Plant Sci 3:167
Grabov A (2007) Plant KT/KUP/HAK potassium transporters: single family—multiple functions. Ann Bot London 99:1035–1041
Greiner T, Ramos J, Alvarez MC, Gurnon JR, Kang M, Van Etten JL, Moroni A, Thiel G (2011) Functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant J 68:977–986
Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q (2008) KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genom 280:437–452
Han Y, Yin S, Huang L (2015) Towards plant salinity tolerance-implications from ion transporters and biochemical regulation. Plant Growth Regul 76:13–23
Han M, Wu W, Wu WH, Wang Y (2016) Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol Plant 9:437–446
He C, Cui K, Duan A, Zeng Y, Zhang J (2012) Genome-wide and molecular evolution analysis of the poplar KT/HAK/KUP potassium transporter gene family. Ecol Evol 2:1996–2004
Higo K, Ugawa Y, Iwamoto M, Higo H (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26:358–359
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:357–359
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Hyun TK, Rim Y, Kim E, Kim JS (2014) Genome-wide and molecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genom 36:365–374
Jack DL, Paulsen IT, Saier MH (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814
Kim EJ, Kwak JM, Uozumi N, Schroeder JI (1998) AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10:51–62
Kobayashi D, Uozumi N, Hisamatsu SI, Yamagami M (2010) AtKUP/HAK/KT9, a K+ transporter from Arabidopsis thaliana, mediates Cs+ uptake in Escherichia coli. Biosci Biotechnol Biochem 74:203–205
Li W, Xu G, Abdel A, Ling Y (2017) Plant HAK/KUP/KT K+ transporters: function and regulation. Semin Cell Dev Biol
Livak KJ, Schmittgen TD (2012) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25:402–408
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16:510–519
Mäser P, Gierth M, Schroeder JI (2002) Molecular mechanisms of potassium and sodium uptake in plants. Plant Soil 247:43–54
Nieves-Cordones M, Ródenas R, Chavanieu A, Rivero RM, Martinez V, Gaillard I, Rubio F (2016) Uneven HAK/KUP/KT protein diversity among Angiosperms: species distribution and perspectives. Front Plant Sci 7:127
Osakabe Y, Arinaga N, Umezawa T, Katsura S, Nagamachi K, Tanaka H et al (2013) Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25:609–624
Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59:595–607
Qiao X, Li M, Li L, Yin H, Wu J, Zhang S (2015) Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol 15:12
Rehman HM, Nawaz MA, Shah ZH, Daur I, Khatoon S, Yang SH et al (2017) In-depth genomic and transcriptomic analysis of five K+ transporter gene families in soybean confirm their differential expression for nodulation. Front Plant Sci 8:804
Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A et al (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13:139–151
Rogiers SY, Coetzee ZA, Walker RR, Deloire A, Tyerman SD (2017) Potassium in the grape (Vitis vinifera L.) berry: transport and function. Front Plant Sci 88:1629
Rubio F, Santa-María GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109:34–43
Santa-María GE, Rubio F, Dubcovsky J, RodrÃguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9:2281–2289
Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A (2001) Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J Biol Chem 276:44563–44569
Shen Y, Shen L, Shen Z, Jing W, Ge H, Zhao J, Zhang W (2016) The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ 38:2766–2779
Shen C, Wang J, Jin X, Liu N, Fan X, Dong C, Shen Q, Xu Y (2017) Potassium enhances the sugar assimilation in leaves and fruit by regulating the expression of key genes involved in sugar metabolism of Asian pears. Plant Growth Regul 1:1–14
Song Z, Guo S, Zhang C, Zhang B, Ma R, Korir NK, Yu M (2015a) KT/HAK/KUP potassium transporter genes differentially expressed during fruit development, ripening, and postharvest shelf-life of ‘Xiahui6’ peaches. Acta Physiol Plant 37:131
Song ZZ, Yang Y, Ma RJ, Xu JL, Yu ML (2015b) Transcription of potassium transporter genes of KT/HAK/KUP family in peach seedlings and responses to abiotic stresses. Biol Plant 59:65–73
Thiel T, Graner A, Waugh R, Grosse I, Close TJ, Stein N (2009) Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol Biol 9:209
Véry AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54:575–603
Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40:523–535
Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64:451–476
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wang Q, Guan C, Wang P, Lv ML, Ma Q, Wu GQ et al (2015) Athkt1;1 and athak5 mediate low-affinity Na+ uptake in Arabidopsis thaliana under mild salt stress. Plant Growth Regul 75:615–623
Wang Y, Liu H, Wang S, Li H (2017) Genome-wide identification and expression analysis of the YUCCA gene family in soybean (Glycine max L.). Plant Growth Regul 81:265–275
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xu Y, Hui L, Li X, Jing L, Wang Z, Yang Q, Chang Y (2015) Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol Plant 37:40
Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G et al (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166:945–959
Zhang Z, Zhang J, Chen Y, Li R, Wang H, Wei J (2012) Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol Biol Rep 39:8465–8473
Zhou H, Qi K, Liu X, Yin H, Wang P, Chen J, Wu J, Zhang S (2016) Genome-wide identification and comparative analysis of the cation proton antiporters family in pear and four other Rosaceae species. Mol Genet Genom 291:1727–1742
Zörb C, Senbayram M, Peiter E (2014) Potassium in agriculture: status and perspectives. J Plant Physiol 171:656–669
Acknowledgements
This work was supported by an award from the China Agriculture Research System (CARS-28-10).
Author information
Authors and Affiliations
Contributions
YX, CD and QS conceived and designed the experiments; YL, LP and CX performed the experiments; YL and XS analysed the data; YL contributed reagents/materials/analysis tools; YL wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have conflict of interest. All authors read and approved the final manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
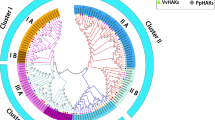
Phylogenetic tree showing HAK/KUP/KTs from Arabidopsis thaliana, Oryza sativa, and Pyrus bretschneideri. The tree was generated with Clustal X2.0 using the NJ method with MEGA6.0. Supplementary material 1 (TIF 1263 KB)
Fig. S2
Phylogenetic tree and PbHAK gene structures. The phylogenetic tree on the left side was generated by MEGA 6.0 using the NJ method with 1000 bootstrap replicates. The percentage bootstrap scores are indicated on the nodes. The exon and intron organization for each PbHAK gene was obtained from the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Exons and introns are represented by pink boxes and black lines, and untranslated regions (UTRs) are indicated by blue boxes. Supplementary material 2 (TIF 698 KB)
Fig. S3
Distribution of conserved motifs within PbHAKs. The phylogenetic tree on the left side was generated by MEGA 6.0 using the NJ method with 1000 bootstrap replicates. Motifs are represented by colored boxes. Motifs were detected using MEME (http://meme-suite.org/tools/meme) with the following parameters: 5 ≤ optimum motif width ≤ 200; the number of motifs = 15; zero or one occurrence per sequence. Supplementary material 3 (TIF 658 KB)
Fig. S4
Chromosomal localization of PbHAK genes. Each bar represents a chromosome, and chromosome numbers are shown at the top of each chromosome. Tandem duplications are shown using red text. The black lines connect segmentally duplicated gene pairs, and the segmental duplication regions were estimated using the Plant Genome Duplication Database (PGDD). Additionally, one pair of segmentally duplicated genes (PbHAK4/PbHAK5) was located on Chr 07 and the other pair (PbHAK10/PbHAK11) was located on Chr 11. Supplementary material 4 (TIF 1187 KB)
Rights and permissions
About this article
Cite this article
Li, Y., Peng, L., Xie, C. et al. Genome-wide identification, characterization, and expression analyses of the HAK/KUP/KT potassium transporter gene family reveals their involvement in K+ deficient and abiotic stress responses in pear rootstock seedlings. Plant Growth Regul 85, 187–198 (2018). https://doi.org/10.1007/s10725-018-0382-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0382-8