Abstract
Sideritis gulendamii is a perennial, endangered plant endemic to Türkiye with narrow and fragmented populations encountered in gypsum and marl soil. This species is known as mountain tea and also has a significant potential for use for medicinal and aromatic purposes. Assessing the genetic diversity of this species holds significant importance in its conservation efforts due to its specific habitat requirements, small and isolated populations, and recent threats imposed on these populations. This study determined genetic diversity at the species and population levels using the inter simple sequence repeat (ISSR) fingerprinting method on 157 individuals from seven natural populations. We observed that the species-level genetic diversity is relatively high (H = 0.189, I = 0.298), but the population-level is low (H = 0.150, I = 0.231). The greater total genetic variation within populations was determined as 76% using the analysis of molecular variance (AMOVA). The obtained value of genetic differentiation (Gst = 0.208, Φpt = 0.24) among populations is considered to be at a moderate level, whereas gene flow appears to be high (Nm = 1.899). Supportive results were reached using the STRUCTURE cluster analysis, Unweighted Pair Group Method with Arithmetic Averages (UPGMA) dendrogram, and Principal Coordinate Analysis (PCoA). The positive correlation between the matrices of genetic distance and geographic distance is determined to be statistically significant, with values of r = 0.566 and p < 0.001 in the Mantel test. Some strategies were developed using the genetic variability pattern of this endangered species for its conservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are 11,707 plant taxa in Türkiye and 3,649 of them are endemic (Güner et al. 2012), so Türkiye can be recognized as a globally significant biodiversity center. Within this rich biodiversity, medicinal and aromatic plants play a significant role in terms of their usage and export (Samet and Cikili 2015). Ethnobotanical research has recorded 1546 species as medicinal plants in Türkiye (Tuzlacı, 2016). In Türkiye, approximately 350 plants collected from nature are traded, and of these plants, about 100 are exported (Akalın et al. 2020). The Lamiaceae family includes many genera collected from nature for medicinal purposes (Samet and Cikili 2015). Sideritis L. is one of these genera, and it is often consumed for preparing herbal tea and traditional folk medicine (Bertsouklis et al. 2022). In addition, folk medicine reports anti-inflammatory, spasmolytic, antiulcer, analgesic, and gastroprotective properties (Tadić et al. 2012, Menković et al. 2013).
Among the Lamiaceae family's largest genera, Sideritis L. is comprises of large number of species over 142 (WCSP 2023). These species, having global economic importance, grow as annual or perennial aromatic herbs (or shrubs) (Chrysargyris et al. 2023). The majority of the species are located in the Mediterranean region, with Spain and Türkiye having the highest number of species. Most of these species are found in the southeastern part of the Iberian Peninsula and the Canary Islands in Spain, with the Marmara and Aegean regions prevailing in Türkiye (González-Burgos et al. 2011). Considerable amount of Sideritis species worldwide are found in Türkiye with 61 taxa; 41 of which are endemic (endemism rate 67.21%) (Güner et al. 2012). Such a high endemism rate is explained by Türkiye’s being an important gene center for this genus with the Iberian Peninsula in southern Europe is the other important gene center (Baser 2002). The genus Sideritis is commonly known as mountain tea in Türkiye (Güner et al. 2012).
Sideritis gulendamii H. Duman & Karavel. is a perennial species with 20–60 cm simple or branched stems, covered with densely adpressed white wooly tomentose hairs, sometimes glabrescent above and eglandular (Duman et al. 1995). It has a chemical structure similar to the traditionally used herbs, hence has potential for medicinal and aromatic purposes (Kirimer et al. 2000, Azizoglu et al. 2011, Çarıkçı et al. 2023). The IUCN Red List Criteria classifies it as Endangered (EN) (IUCN 2022). It has narrow and isolated seven small natural populations in the Central Anatolia in Türkiye (Ankara and Eskişehir provinces). It has specific habitat requirements and grows only over gypsum and marly soils. Most importantly, this species' populations are at risk due to various factors such as the expansion of agricultural areas, overgrazing, proximity to both highways and urban areas, reforestation, terracing, and erosion (Yıldırım et al. 2019). As a result, for these species effective conservation measures must be taken.
Understanding the degree of genetic diversity, especially in endemic and endangered species, is crucial as it forms the foundation for crafting effective conservation strategies. Molecular markers might reveal details about genetic variation and adaptive evolutionary processes (Garayalde et al. 2011). Molecular marker techniques are widely used in plant sciences to evaluate ecological, evolutionary, taxonomic, phylogenetic, and genetic research. Generally, the choice of a molecular marker must strike a balance among several parameters such as dependability, confidence in showing polymorphisms, statistical power, and ease of analysis (Agarwal et al. 2008). Inter simple sequence repeats (ISSR) is one of the commonly used molecular markers in research regarding population genetics, particularly for plants (Gupta et al. 1994, Li and Jin 2008). ISSR represents a quick, simple, and highly repeatable method. ISSR markers typically exhibit high polymorphism, are cost-effective, and their key advantage lies in not requiring prior knowledge of the genomic sequence (Zietkiewicz et al. 1994, Bornet and Branchard 2001, Idrees and Irshad 2014). It is aimed to investigate the genetic diversity and differentiation patterns of seven small natural populations of S. gulendamii using ISSR markers for developing effective conservation strategies.
Materials and methods
Plant material
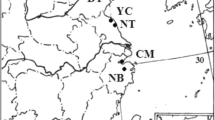
Field experiments on the S. gulendamii species were carried out in the Central Anatolian region's provinces of Ankara and Eskişehir in July and August of 2016. 157 fresh leaf samples were gathered from the known seven natural populations of the species (Fig. 1) (Table 1). The sampling method depended on the size of each population, and care was taken to choose the sampled individuals as far apart as possible. Fresh leaf samples collected during the fieldwork were stored and dried in sealed bags containing silica gel. The dried leaf samples were maintained at –20 °C until use.
DNA extraction and ISSR-PCR amplification
Utilizing established protocols of the Macherey–Nagel NucleoSpin Plant II kit, dried leaf tissues yielded good quality genomic DNA samples. After determining the quality and quantity of DNA samples by using the Nanodrop spectrophotometer, the samples were diluted to a concentration of 10 ng/μl and stored at –20 °C until PCR process. During the amplification process, 36 ISSR primers were screened, and from these, the top 10 primers were chosen depending on repeatability, polymorphism rate, and clear banding patterns suitable for the study (set no. 9, University of British Columbia 2006). PCR applications were carried out using "Applied Biosystems Veriti Thermal Cycler" and a touch down PCR (TD) program was used (Don et al. 1991). The ISSR-PCR reaction components are listed in Table 2 with their volumes and concentrations. The ISSR amplification conditions comprised an initial denaturation step for 5 min at 95 °C, followed by 1 cycle; denaturation for 30 s at 95 °C, annealing for 45 s at 65–55 °C, extension for 1 min 30 s at 72 °C, followed by 15 cycles; denaturation for 30 s at 95 °C, annealing for 30 s at 45 °C, extension for 1 min 30 s at 72 °C, followed by 20 cycles; and a final extension for 7 min at 72 °C, followed by 1 cycle. Then, these products were electrophoresed on a 2% agarose gel containing 0.5X TBE buffer (Tris–Borate-EDTA) and ethidium bromide (EB) solution for approximately 5 h at a voltage of 90 V. The sizes of the amplified DNA fragments were determined by comparing them to the ABM 100 bp ladder. The stained gel was visualized under UV light using the 'BioRad Molecular Imager DocXR + '.
Data analysis
A binary data matrix was constructed for statistical evaluation based on the amplified fragment presence ‘1’ or absence '0'. The genetic diversity parameters, which include Shannon’s information index (I) (Lewontin 1972), Nei’s gene diversity (H) (Nei 1973), number of observed and effective alleles (Na and Ne, respectively) (Kimura and Crow 1964), number and percentage of polymorphic loci (NPL and PPL, respectively), genetic diversity within population (Hs), total genetic diversity (Ht), and coefficient of genetic differentiation among populations (Gst = 1–Hs/Ht), were obtained from POPGENE v1.32 (Yeh et al. 1997). Gene flow (Nm) was extracted using the Gst values in Nm = 0.5(1–Gst)/Gst (McDermott and McDonald 1993). The values of polymorphism information content (PIC) were obtained by using the frequency of the amplified allele (ƒi) values in PICi = 2ƒi (1–ƒi) (Abuzayed et al. 2017).
Molecular Evolutionary Genetics Analysis (MEGA) 6.06 software (Tamura et al. 2013) was used for depicting an Unweighted Pair Group Method with Arithmetic Mean (UPGMA) dendrogram showing the genetic relationship between populations based on the matrix of Nei's genetic distances (Nei 1978). SYN-TAX 2000 software was used for depicting another UPGMA dendrogram utilizing the Jaccard similarity coefficient (Jaccard 1908, Podani 2001) to reflect the genetic distances between individuals. A Principal Coordinate Analysis (PCoA) (Orlóci 1978) was carried out on the basis of Nei's genetic distances (Nei 1978) using GenAlEx 6.5 software to support the clustering patterns formed by the UPGMA dendrograms (Peakall and Smouse 2012). Applying the Analysis of Molecular Variance (AMOVA) in GenAlEx 6.5, the genetic variation was assessed both within and among populations (Excoffier et al. 1992). In order to assess the genetic differentiation among populations, the variation was summarized with the Φpt statistic, an analogue of Fixation index (Fst). The significance test was calculated based on 999 permutations. The Mantel test was conducted using GenAlEx 6.5 for the evaluation of the relationship between geographic and genetic distance matrices, employing 999 permutations for testing (Mantel 1967).
A Bayesian clustering method was utilized in STRUCTURE v 2.3.4 software for investigating the genetic structure of S. gulendamii populations (Pritchard et al. 2000). Employing Markov chain Monte Carlo repetitions of 300,000 and a burn-in period of 50,000 iterations, 20 independent runs of K (ranging from 2 to 20) were conducted utilizing an admixture model to find out the best number of clusters. With the help of STRUCTURE Harvester software (Earl and VonHoldt 2012), the best-fit value of K was obtained by evaluating the ΔK statistic and the Ln probability of data, LnP(K) (Evanno et al. 2005). The threshold value of ≥ 0.80 was used to assign populations to clusters (A, B, C, and D). Individuals were regarded as admixed when they did not meet this threshold (Celik et al. 2016).
Results
The analysis of the selected 10 ISSR primers for the S. gulendamii species resulted in a total of 236 bands from 157 individuals representing seven populations, out of which 226 bands were polymorphic (95.8%) (Table 3). Among the primers used, UBC 807 and 810 were the primers with the largest polymorphism ratio (100%), whereas UBC 903 was the primer with the smallest polymorphism ratio (90%). Across all samples, the number of amplified bands per primer ranged from 18 to 34 with an average of 23.6 whereas the average for polymorphic bands was 22.6. The primer UBC 888 yielded the greatest number of bands in both total bands and polymorphic bands. The primer UBC 890 produced the least number of bands in both total and polymorphic bands. The size range of PCR fragments was between 200 and 2000 bp. PIC values were obtained between 0.174 and 0.266 and mean value for PIC was found to be 0.200. A PIC value greater than 0.50 generally suggests that the markers were very informative (Botstein et al. 1980, Serrote et al. 2020) therefore this study’s PIC values can be considered to be moderately informative.
The ISSR data were analyzed using POPGENE 1.32 for the assessment of genetic diversity and differentiation (Tables 4 and 5). The ISSR analysis revealed that the number of observed alleles (Na) were between 1.495 and 1.601, with an average of 1.541; the number of effective alleles (Ne) were between 1.227 and 1.257, with an average of 1.249; The Nei's gene diversity (H) was between 0.137 and 0.157, with an average of 0.189; The Shannon's information index (I) was between 0.212 and 0.246, with an average of 0.298. Furthermore, the percentage of polymorphic loci (PPL) was between 49.6% and 60.2%, with an average of 54.2% (Table 4).
The total gene diversity (Ht) from all loci data for the species was calculated as 0.189, while the average gene diversity within population (Hs) was found to be 0.150. The coefficient of differentiation among populations (Gst) was 0.208, which indicates that approximately 20% of total genetic variation occurred among populations and the remaining 80% within populations. Gene flow (Nm) was considered to be high when it was larger than 1 and low when it was smaller than 0.249, while it was considered to be intermediate between these values (Slatkin 1981, 1985, Caccone 1985, Waples 1987). The gene flow (Nm) of S. gulendamii was determined to be 1.899, suggesting that the genetic migration among populations was at high levels. (Table 5). The AMOVA results highlighted striking genetic variation within populations (76% of the total variation) compared to the genetic variation among populations (24% of the total variation). The total differentiation among populations (Φpt) was obtained as 0.24 (Table 6). This analysis, which was based on 999 iterations of permutations, was regarded statistically significant (p < 0.001). Genetic differentiation is said to be low, moderate, high and very high depending on the Φpt value; 0–0.05, 0.05 −0.15, 0.15–0.25, and above 0.25, respectively (Wright 1978, Hartl and Clark 1997). In a related context, obtained value of Φpt (0.24) pointed out a moderate genetic differentiation existed among populations.
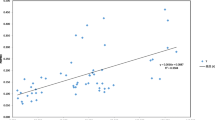
Nei’s unbiased genetic distance varied from 0.019 to 0.080 between seven populations of S. gulendamii (Nei 1978) (Table 7). The smallest genetic distance (0.019) was determined among the populations of YD and CR. These two populations are also geographically only 15 km apart from each other. The highest genetic distance (0.080) was determined among the populations of AK and IN. The geographic distance separating these two populations is 100 km. The positive correlation between the matrices of genetic distance and geographic distance was determined to be statistically significant with values of r = 0.566 and p < 0.001 in Mantel test (Fig. 2).
Employing Nei's binary genetic distance matrix (Nei 1978), the relationship among S. gulendamii populations was investigated by UPGMA and PCoA cluster analyses. The UPGMA dendrogram distinctly separated the clustered 7 populations into two main groups, representing populations found in Ankara and Eskişehir provinces (Figs. 3, 4). While the Ankara province group was composed of BS, IN and SR populations and connected AY population, the Eskişehir province group was composed of CR and YD, and connected AK population (Fig. 3). A similar clustering was also observed in the PCoA plot. The PCoA plot based on populations results revealed that 45.6%, 18.3%, and 15.1% of the total variance was explained by axis 1, axis 2, and axis 3, respectively (Fig. 4).
For a detailed visualization of the genetic relationships between genotypes of S. gulendamii, we performed both UPGMA and PCoA clustering analyses (Each population is shown by a different color) (Figs. 5, 6). In the UPGMA dendrogram, individuals typically showed a tendency to cluster together within their respective populations. In detail, Ankara populations, the first main cluster at UPGMA dendogram, were composed of Group A1, the subgroups A1-1, A1-2, and A1-3. While the subgroup A1-1 consisted of the all genotypes of AY population and one genotype of BS population, the subgroup A1-2 consisted of the genotypes of BS and IN which clustered together and one genotype of SR population. Sub-group A1-3 was completely clustered with the genotypes of SR population. The second main cluster, Eskişehir populations, consisted of Group B1, and the subgroups B1-1 and B1-2. While the subgroup B1-1 consisted of CR and YD populations that clustered together, the sub-group B1-2 consisted of the genotypes of AK population (Fig. 5). The PCoA plot at the individual genotype level supports the UPGMA dendrogram. It indicates that axis 1, axis 2, and axis 3 accounted for 15.03%, 6.83%, and 6.14% of the total variance (Fig. 6).
According to the STRUCTURE analysis conducted for K = 4, genotypes of S. gulendamii were assigned to subclusters (A, B, C, and D) if their probability of membership (threshold) value qi > 0.80. Admixed cluster did not meet criteria; i.e. the qi value is smaller than 0.80 for this cluster (See supplementary information). Cluster A comprised 21 genotypes (13.3%) solely from the SR population. Cluster B consisted of 42 genotypes (26.7%), comprising 19 and 23 genotypes from the BS and IN populations, respectively. Having 66 genotypes (42.03%), cluster C had the largest number of genotypes. This cluster contains 22, 21 and 23 genotypes from the populations of CR, YD, and AK, respectively. Cluster D included 21 genotypes (13.3%) exclusively from the AY population. Additionally, an admixed cluster was composed of 7 genotypes (4.45%) from all populations except the IN population (Fig. 7).
STRUCTURE analysis of Sideritis gulendamii based on ISSR data for 157 individuals (7 populations). a The variation of delta K vs. K values for the estimation of the number of cluster populations. b The output table according to the Evanno method highlighting the highest ΔK value at K = 4. c The STRUCTURE output of the analysis for K = 4 clusters. The seven populations were seperated by vertical black lines. The same color represents the same cluster. (Color figure online)
Discussion
It has been estimated that 31.82% of plant taxa are endemic for Türkiye (Ekim et al. 2000, IUCN 2022). The majority of these taxa are in the categories of CR, EN, and VU due to high risk while a tiny portion of them (0.03%) falls into EX (Türe and Böcük 2010). Therefore, it is very important to conduct research on plant species that need conservation. Hence, this work aimed to determine the genetic diversity of the natural populations of S. gulendamii, which is know to be endangered and confined to only gypsum and marly soils in Central Anatolia.
The percentage of polymorphic locus in Sideritis gulendamii was observed to be 95.8% (PPLsp) at the species level, while it was found to be 54.2% (PPLpop) at the population level. Research on other Lamiaceae species using ISSR markers have shown varying levels of polymorphism in terms of the percentage of polymorphic locus (PPL). For instance, wide spread endemic species Lamiophlomis rotate (Benth.) Kudo (PPLsp = 96.73%) (Liu et al. 2006), an endangered medicinal and aromatic species Cunila spicata Benth. (PPLsp = 84.4%, PPLpop = 13.94%) (Echeverrigaray et al. 2016), narrowly distributed, endemic and threatened species Satureja khuzistanica Jamzad (PPLsp = 98.33%, PPLpop = 74.88%) (Hadian et al. 2017), Teucrium leucophyllum Montbret & Aucher ex Benth. (PPLsp = 99.31%, PPBpop = 79.14%) (Sözen et al. 2017), Scutellaria yildirimlii Çiçek & Yaprak (PPLsp = 93.9%, PPLpop = 56.5%) (Yıldırım et al. 2023). Primula cicutariifolia Pax (PPLsp = 64.8%, PPLpop = 25.7%) (Wang et al. 2014), Verbascum gypsicola Vural & Aydoğdu (PPLsp = 99.5%, PPLpop = 61.1%) (Keser and Yaprak 2023), Muscari adilii M. B. Güner & H. Duman (PPLsp = 97.0%, PPLpop = 69.6%) (Keser et al. 2023) are the examples for narrow endemic and endangered species from other families. The percentage of polymorphic locus ranged from 64.8% and 99.5% at the species level, and from 13.94 and 79.14% at the population level. The PPL values of S. gulendamii indicate a high polymorphism rate at the species level but moderate polymorphism rate at the population level.
Shannon's information index (I) and Nei's gene diversity (H) of S. gulendamii were calculated at the species level (Isp = 0.298, Hsp = 0.189) and population level (Ipop = 0.231, Hpop = 0.150). Values corresponding to narrowly distributed endemic and endangered species from the Lamiaceae were Scutellaria yildirimlii Çiçek & Yaprak (Isp = 0.292, Hsp = 0.183 and Ipop = 0.242, Hpop = 0.158) (Yıldırım et al. 2023), Satureja khuzistanica Jamzad (Isp = 0.466, Hsp = 0.306 and Ipop = 0.390, Hpop = 0.262) (Hadian et al. 2017), Teucrium leucophyllum Montbret & Aucher ex Benth. (Isp = 0.418, Hsp = 0.263 and Ipop = 0.345, Hpop = 0.220) (Sözen et al. 2017), from other families Uechtritzia armena Freyn & Sint. (Isp = 0.171, Hsp = 0.192 and Ipop = 0.275, Hpop = 0.333,) (Yıldırım Doğan et al. 2016), Verbascum gypsicola Vural & Aydoğdu (Isp = 0.25, Hsp = 0.15, and Ipop = 0.21, Hpop = 0.13) (Keser and Yaprak 2023), and Muscari adilii M.B.Güner & H.Duman (Isp = 0.300 Hsp = 0.188 and, Ipop = 0.268, Hpop = 0.171) (Keser et al. 2023). Comparing these results with those of S. gulendamii, we observed that the genetic diversity at species level is relatively high while that at population level is low. As generally expected, the genetic diversity of endemic and locally distributed species is low (Hamrick and Godt 1996). Nevertheles, the genetic diversity of S. gulendamii is obtained at high levels at the species level. This result can be explained by Türkiye’s being one of the primary gene hubs for the Sideritis genus.
If we were to rank the populations of S. gulendamii based on genetic diversity values (H and I), the order from the highest to lowest would be YD, CR, SR, AY, AK, IN and BS. On the other hand, one of the main factor influencing genetic diversity levels is the population size. In the study by Yıldırım et al. (2019), these populations were ranked from largest to smallest as AK, IN, SR, CR, BS, YD, and AY based on their population sizes. Although the AK (8338 individuals) and IN (4244 individuals) populations have the highest number of individuals, they are at the fifth and sixth place, respectively, in terms of genetic diversity. Two explanations can clarify this result; the number of individuals decreased greatly in the past and then increased again, or the settlement of new individuals of this species in the area and the founder effect. Genetic diversity was found to be proportional to population size for SR population (3559 individuals). The distribution area of this population is located at approximately 1648 m and is more isolated than the other populations away from various anthropogenic influences. It is thought that the number of individuals in this population did not show great changes in the past, and there was no stochastic effect. Although the BS population was represented by 1285 individuals, it has the lowest genetic diversity. This population is very close to settlements which suggests that there were fluctuations in population size in the past. YD population which has only 884 individuals, has the highest genetic diversity that can be explained with its proximity (15 km) to CR population with 1743 individuals and probable gene flow. The AY population, which has the smallest number of individuals, is at the forth place out of seven populations in terms of genetic diversity. This population was larger until recently but has shown a sudden shrinkage since 2010 due to field clearing, afforestation, area destruction and habitat loss (Yıldırım et al. 2023). While the possible distribution area for AY population is much wider, the fact that 510 individuals were confined to an area of 2.2 ha can be shown as evidence to explain this situation.
The genetic differentiation among populations is shaped by various factors, including habitat fragmentation, selection, gene flow, mating patterns, and genetic drift (Schaal et al. 1998). These also could be linked to the total amount of genetic variation and its distribution within and among populations. The distribution of genetic diversity in plant species is usually associated with their breeding mechanisms (Hamrick and Godt 1996). The long-lived perennial species exhibiting outcrossing tend to keep the majority of the genetic diversity within populations, whereas annual species exhibiting selfing tend to distribute majority of the genetic diversity among populations. Average coefficients reported for long-lived perennials are Φpt = 0.25 and Gst = 0.19 whereas they are Φpt = 0.27 and Gst = 0.22 for outcrossing species (Nybom 2004). Among the populations of S. gulendamii, the genetic differentiation values were obtained as Φpt = 0.24 and Gst = 0.208, consistent with findings in outcrossing plant species. The AMOVA test results indicated that 76% of the total genetic variation is due to differences among individuals within populations while 24% of it is due to population differences. High value of genetic variation within the populations of S. gulendamii is likely because of the outcrossing breeding mechanism of the species.
Papaporfyriou et al. (2020) draw attention to a significant decline in the populations of Sideritis scardica (Greek sage), an endemic medicinal and vulnerable plant species of Central Balkan Peninsula, which is also under the influence of the same anthropogenic factors as Sideritis gulendamii. The biotic factors, abiotic factors, and anthropogenic factors have very crucial role on the survival of the species in long term. In addition, a previous molecular study with AFLP markers on Sideritis species reported that a statistically significant and relatively high genetic variation (Fst = 0.344, p < 0.0001) between the populations of S. euboea (Greece) exists (Sarrou et al. 2022). Similarly, noticable genetic differentiation (Φpt = 0.367, p < 0.0001) among populations of S. scardica Griseb. (Balkan Peninsula) was observed using the AFLP marker (Grdiša et al. 2019). However, in contrast, the use of URP molecular markers found high rates of population differentiation of endemic S. raeseri Boiss. & Heldr. (Balkan Peninsula) (Φpt = 0.633, p < 001) (Patelou et al. 2020). The genetic differentiation among the population of S. gulendamii was statistically significant at a moderate level (Φpt = 0.24, p < 0.001), consistent with the obtained value of Gst (0.208), but it was lower than in previous research as mentioned above. Gene flow is a major factor affecting the genetic structure. Gene flow value of S. gulendamii was partially high level (Nm = 1.899). When Nm value is larger than 1, the gene flow is expected to be at high levels and this can hinder genetic differentiation among populations resulting from genetic drift (Slatkin and Barton 1989, Yan et al. 2019). The dispersal of seeds or pollens by long distances is believed to be the reason for the high value for gene flow (Sözen et al. 2017). Pollination through insects and birds, and cross-pollination are widespread in the Lamiaceae family (Watson and Dalwitz 1992, Judd et al. 2015). The individuals within this family provides pollen and nectar bees. In addition, it is thought that the Macroglassum stellatarum (Sphingidae) species, which was observed to visit the flowers of the S. gulendamii species during field research, make significant contribution in shaping the genetic structure of the species by carrying out pollen transfer. Moderate genetic differentiation and high gene flow were observed in insect-pollinated steppe perennials, like Teucrium leucophyllum (Sözen et al. 2017) and Scutellaria yildirimlii (Yıldırım et al. 2023). Both are endemic for Türkiye that exhibit a narrow distribution and are categorized as endangered (EN). Besides, Scutellaria yildirimlii and Sideritis gulendamii are distributed together in AY and AK localities. According to Yıldırım et al. (2023), this situation could be the result of different natural selection processes due to the small population size.
According to the STRUCTURE clustering analysis, the seven populations of S. gulendamii have been separated into four distinct genetic groups. The first cluster consists of AY; the second one includes BS and IN; the third one comprises SR; and the fourth one is composed of CR, YD, and AK. The separation has been supported both by the UPGMA dendrogram and the PCoA plot. Based on the Mantel test, a statistically significant positive correlation (r = 0.566, p < 0.001) was found between geographical and genetic distances. Mantel test results are supported by the UPGMA and PCoA cluster analyses. In this case, it can be said that geographic distance also contributes to genetic differentiation among populations. In both the UPGMA dendrogram and PCoA clustering analyses, Ankara populations (AY, BS, IN and SR) and Eskişehir populations (CR, YD, and AK) were clearly separated from each other. Regardless of the type of the analysis, the BS population was clustered with the geographically closest population (5 km), IN population. The genetic distance was measured as 0.029. Despite of the 15 km geographically separated each other, CR and YD populations were found to be two genetically closest populations. In addition, the lowest genetic distance value of 0.019 was obtained for these two populations. UPGMA dendrogram in Fig. 5 shows that the populations are clearly separated from each other, with a few exceptions. An individual (number 46) from BS population clustered with AY population which is 48 km distance apart, and an individual (number 71) from SR population clustered with IN population which is 60 km distance apart. This situation, also seen in S. yildirimlii, suggests the possibility of long-distance pollen or seed distribution between populations (Yıldırım et al. 2023).
Conclusion
Sideritis gulendamii is an endemic plant species that has seven small, narrowly distributed and isolated populations with special habitat requirements due to edaphic factors. In situ conservation of natural populations is a priority step for maintaining this species’ genetic diversity. This is especially important for populations like YD, CR, and AY, which have low numbers of individuals but high genetic diversity. These populations should be given conservation priorities, because individuals of these populations are likely to be more competitive than the individuals of populations with low genetic diversity and high numbers of individuals. These small but genetically diverse populations have higher ecological plasticity and can more easily adapt themselves to changing environmental conditions. Besides, anthropogenic factors such as overgrazing, reforestation, expansion of agricultural areas, and field clearings put pressure on this species. In order to prevent these anthropogenic effects, there should be some cooperation between relevant institutions and organizations. To provide the continuation of gene flow between populations, prevention of habitat fragmentation and habitat loss is very important for in situ conservation measures. Ex-situ conservation research can be carried out. From all known populations, seed samples can be collected and preserved to be as representative as possible of genetic diversity. Since many populations of the species are close to settlements, they are easily accessible, and it is important to inform the local people about this issue in order not to collect too much of this species, which can be consumed as tea.
Briefly, comprehending the population structure and genetic diversity of plant species is crucial for effective conservation efforts. The findings of this study indicates that, as a powerful and useful tool, ISSR molecular markers can successfully be employed for assessing the genetic structure of the endemic and endangered S. gulendamii.
References
Abuzayed M, El-Dabba N, Frary A, Doganlar S (2017) GDdom: an online tool for calculation of dominant marker gene diversity. Biochem Genet 55:155–157
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 27:617–631
Akalın E, Gürdal B, Olcay B (2020) General overview on the conservation of medicinal plants in Turkey. Turkish J Biodivers 3:86–94
Azizoglu A, Özer Z, Kilic T (2011) An experimental and theoretical study on siderol isolated from Sideritis species. Collect Czechoslov Chem Commun 76:95–114
Baser KHC (2002) Aromatic biodiversity among the flowering plant taxa of Turkey. Pure Appl Chem 74:527–545
Bertsouklis K, Theodorou P, Aretaki P-E (2022) In vitro propagation of the mount parnitha endangered species Sideritis raeseri subsp, Attica. Horticulturae 8:1114
Bornet B, Branchard M (2001) Nonanchored inter simple sequence repeat (ISSR) markers: reproducible and specific tools for genome fingerprinting. Plant Mol Biol Report 19:209–215
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Caccone A (1985) Gene flow in cave arthropods: a qualitative and quantitative approach. Evolution 39:1223–1235
Çarıkçı S, Kılıç T, Gören AC et al (2023) Chemical profile of the Anatolian Sideritis species with bioactivity studies. Pharm Biol 61:1484–1511
Celik I, Camci H, Kose A et al (2016) Molecular genetic diversity and association mapping of morphine content and agronomic traits in Turkish opium poppy (Papaver somniferum) germplasm. Mol Breed 36:1–13
Chrysargyris A, Tomou E-M, Goula K et al (2023) Sideritis L. essential oils: a systematic review. Phytochemistry 209:113607
Don RH, Cox PT, Wainwright BJ et al (1991) ‘Touchdown’PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Duman H, Aytaç Z, Ekici M et al (1995) Three new species (Labiatae) from Turkey. Flora Mediterr 5:221–228
Earl DA, VonHoldt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the evanno method. Conserv Genet Resour 4:359–361
Echeverrigaray S, Albuquerque M, Zacaria J, Longaray AP (2016) Population structure and genetic diversity of the medicinal and aromatic plant cunila spicata benth. (Lamiaceae) based on inter-simple sequence repeat (ISSR) markers. J Adv Biotechnol 5:652–658
Ekim T, Koyuncu M, Vural M, et al (2000) Türkiye bitkileri kirmizi kitabi (Red data book of turkish plants). Türkiye Tabiatını Koruma Derneği, Ankara
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Garayalde AF, Poverene M, Cantamutto M, Carrera AD (2011) Wild sunflower diversity in Argentina revealed by ISSR and SSR markers: an approach for conservation and breeding programmes. Ann Appl Biol 158:305–317
González-Burgos E, Carretero ME, Gómez-Serranillos MP (2011) Sideritis spp.: uses, chemical composition and pharmacological activities—a review. J Ethnopharmacol 135:209–225
Grdiša M, Radosavljević I, Liber Z et al (2019) Divergent selection and genetic structure of Sideritis scardica populations from southern Balkan Peninsula as revealed by AFLP fingerprinting. Sci Rep 9:12767
Güner A, Aslan S, Ekim T, et al (2012) Türkiye bitkileri listesi:(damarlı bitkiler). Nezahat Gökyiǧit Botanik Bahçesi Yayınları
Gupta M, Chyi Y-S, Romero-Severson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor Appl Genet 89:998–1006
Hadian J, Raeisi S, Azizi A et al (2017) Genetic diversity of natural populations of medicinally valuable plant Satureja khuzistanica Jamzad based on ISSR markers. Braz J Bot 40:771–781
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman & Hall, New York, pp 281–304
Hartl DL, Clark AG (1997) Principles of population genetics. Sinauer associates Sunderland
Idrees M, Irshad M (2014) Molecular markers in plants for analysis of genetic diversity: a review. Eur Acad Res 2:1513–1540
IUCN (2022) The IUCN red list categories and criteria, Version 2022-2. https://www.iucnredlist.org. Accessed 20 Nov 2023
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Judd W, Campbell C, Kellogg E, et al (2015) Plant systematics: a phylogenetic approach. Sinauer Associates Sunderland, Massachusetts
Keser AM, Yaprak AE (2023) The assessment of genetic diversity and population structure of Verbascum gypsicola by ISSR markers for conservation purposes. Nord J Bot 2023:e03660
Keser AM, Yaprak AE, Tuğ GN (2023) Determination of the genetic diversity, population structure, and some ecological preferences of the endemic Muscari adilii. Turk J Bot 47:353–362
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Kirimer N, Tabanca N, Özek T et al (2000) Essential oils of annual Sideritis species growing in Turkey. Pharm Biol 38:106–111
Lewontin RC (1972) The apportionment of human diversity. Evolutionary biology. Springer, pp 381–398
Li J-M, Jin Z-X (2008) Genetic structure of endangered Emmenopterys henryi Oliv. based on ISSR polymorphism and implications for its conservation. Genetica 133:227–234
Liu J, Wang L, Geng Y et al (2006) Genetic diversity and population structure of Lamiophlomis rotata (Lamiaceae), an endemic species of Qinghai-Tibet Plateau. Genetica 128:385–394
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McDermott JM, McDonald BA (1993) Gene flow in plant pathosystems. Annu Rev Phytopathol 31:353–373
Menković N, Gođevac D, Savikin K et al (2013) Bioactive compounds of endemic species Sideritis raeseri subsp raeseri grown in national park Galicica. Rec Nat Prod 7:161–168
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Orlóci L (1978) Multivariate analysis in vegetation research, 2nd edn. Springer, Netherlands, The Hague
Papaporfyriou PK, Sarrou E, Avramidou E, Abraham EM (2020) Abundance and phenotypic diversity of the medicinal Sideritis scardica Griseb. In relation to floristic composition of its habitat in northern Greece. Sustainability 12:2542
Patelou E, Chatzopoulou P, Polidoros AN, Mylona PV (2020) Genetic diversity and structure of Sideritis raeseri Boiss. & Heldr (Lamiaceae) wild populations from Balkan Peninsula. J Appl Res Med Aromat Plants 16:100241
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Podani J (2001) SYN-TAX 2000 computer programs for data analysis in ecology and systematics user’s manual. Scientia Publishing, Budapest
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Samet H, Cikili Y (2015) Journal of rural and community development in Tanzania. J Rural Commun Dev 10:75–84
Sarrou E, Doukidou L, Avramidou EV et al (2022) Chemodiversity is closely linked to genetic and environmental diversity: insights into the endangered populations of the local endemic plant Sideritis euboea Heldr. of Evia Island (Greece). J Appl Res Med Aromat Plants 31:100426
Schaal BA, Hayworth DA, Olsen KM et al (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Serrote CML, Reiniger LRS, Silva KB et al (2020) Determining the polymorphism information content of a molecular marker. Gene 726:144175
Slatkin M (1981) Estimating levels of gene flow in natural populations. Genetics 99:323–335
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43:1349–1368
Sözen E, Hilooglu M, Kandemir A (2017) Genetic diversity of local endemic Teucrium leucophyllum Montbret & Aucher ex Bentham. (Lamiaceae) in Turkey. Indian J Pharm Educ Res 51:s195–s199
Tadić V, Jeremic I, Dobric S et al (2012) Anti-inflammatory, gastroprotective, and cytotoxic effects of Sideritis scardica extracts. Planta Med 78:415–427. https://doi.org/10.1055/s-0031-1298172
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Türe C, Böcük H (2010) Distribution patterns of threatened endemic plants in Turkey: a quantitative approach for conservation. J Nat Conserv 18:296–303
Tuzlacı E (2016) Türkiye bitkileri geleneksel ilaç rehberi. İstanbul
Wang D-Y, Chen Y-J, Zhu H-M et al (2014) Highly differentiated populations of the narrow endemic and endangered species Primula cicutariifolia in China, revealed by ISSR and SSR. Biochem Syst Ecol 53:59–68
Waples RS (1987) A multispecies approach to the analysis of gene flow in marine shore fishes. Evolution 41:385–400
Watson L, Dalwitz MJ (1992) The families of flowering plants: descriptions, illustrations, identification, and information Retrieval. Version: 26th October 2019. delta-intkey.com
WCSP (2023) World checklist of selected plant families. Facilitated by the royal botanic gardens, Kew. http://wcsp.science.kew.org/. Accessed 7 Dec 2023
Wright S (1978) Evolution and the genetics of populations. The University of Chicago Press, Chicago
Yan W, Li J, Zheng D et al (2019) Analysis of genetic population structure and diversity in Mallotus oblongifolius using ISSR and SRAP markers. PeerJ 7:e7173
Yeh FC, Yang RC, Boyle TBJ et al (1997) POPGENE, the user-friendly shareware for population genetic analysis. Mol Biol Biotechnol Centre, Univ Alberta, Canada 10:295–301
Yıldırım M, Ayyıldız G, Keser AM et al (2019) Current population sizes, distribution areas and re-evaluated IUCN categories of rare and endemic species from Central Anatolia, Turkey: Salsola grandis, Scutellaria yildirimlii and Sideritis gulendamii. Biyolojik Çeşitlilik Ve Koruma 12:151–160
Yıldırım M, Tuğ GN, Yaprak AE (2023) The assessment of genetic diversity and population structure of endemic Scutellaria yildirimlii (Lamiaceae) for conservation purposes. Turk J Bot 47:211–223
Yıldırım Doğan N, Kandemir A, Osma E (2016) Genetic diversity and variability among populations and ecological characteristics of the Uechtritzia armena Freyn (Asteraceae) endemic to Turkey. 4:20–27
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgements
We express our gratitude for the invaluable assistance provided by Prof. Mecit VURAL, Dr. Gül AYYILDIZ, Dr. Banu AVCIOĞLU, Dr. Ali Murat KESER, and Asst. Prof. İbrahim ÇELİK.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). Funding for this work was provided by Scientific and Technological Research Council of Türkiye (TÜBİTAK) (Grant Number: 117Z425).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work
Corresponding author
Ethics declarations
Conflict of interest
The Authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yıldırım, M., Tuğ, G.N. & Yaprak, A.E. Analyses of genetic diversity and population structure of endemic and endangered species Sideritis gulendamii (Lamiaceae) and implications for its conservation. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01907-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01907-6