Abstract
The genus Astragalus L. with about 3000 species is the largest genus of flowering plants. It is also the largest genus in the flora of Iran with more than 850 taxa. Astragalus cyclophyllon Beck is an endemic taxon belonging to Astragalus sect. Incani DC. It occurs scattered in the steppe and semi-steppe areas of the western and central parts of Iran. This species is one of the most important forage taxa of Astragalus. According to IUCN criteria it has been classified as a vulnerable taxon in Iran, as it occurs in habitats intensely used by herders for their livestock. Here we evaluate the genetic diversity of the populations of this species in Iran to see whether genetic diversity is high or if reductions have already happened. For this purpose, sampling was done in all geographical areas with A. cyclophyllon populations in Iran. In total, 80 individuals representing 29 populations were studied using 33 quantitative and qualitative morphological characters and 10 inter-simple sequence repeat (ISSR) primers. ISSR revealed 240 bands which all were polymorphic. Neighbor-joining cluster analysis divided the individuals in four groups, Principal Coordinate Analysis and Bayesian population assignment analysis in Structure resulted in three genetic units. Morphological variation showed no correlation with the molecular data. The mean of Gst and Nm indices are 0.516 and 0.468, respectively, which indicate a very high genetic differentiation and low gene flow between the studied populations. According to these results, we conclude that genetic diversity is high in this species and that, therefore, the major threat for A. cyclophyllon is currently not related to inbreeding depression in populations, but might be due to livestock grazing that could change the population demographic structure by reducing regular establishment of new offspring.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Astragalus L. belongs to the Fabaceae subfamily Faboideae and is with about 3000 species in more than 250 sections the largest genus in flowering plants (Podlech and Zarre 2013; Maassoumi 2020). It is distributed in the temperate, cold or semi-arid continental regions of Europe, Asia, North America and South America (Podlech 1986). Astragalus has its highest diversity with about 1500 species in the Irano-Turanian floristic region of Southwest Asia, and with more than 850 species it is the largest genus in the flora of Iran (Maassoumi 2020). Astragalus cyclophyllon Beck is grouped in Astragalus sect. Incani DC. It is an endemic taxon to Iran and is scattered in the western and central parts of the country (Maassoumi 2005; Podlech and Zarre 2013). Astragalus cyclophyllon is known to be a valuable forage plant that grows in a large area of rangelands and is effective in preventing soil erosion (Maassoumi 1998). This species has many flowers and a long flowering period. Its nectar is very desirable for the production of honey. In terms of the risk for extinction, it has been rated as a vulnerable species in Iran (Jalili and Jamzad 1999).
DNA-based molecular markers are reliable because they are independent of environmental factors, physiological conditions, and age of the organism (Kalpana et al. 2004). They are the basis of allelic polymorphisms within and among populations. Furthermore, they can provide a large number of characters that are easy to observe, score, and analyze (Lombard et al. 2001; Hasan et al. 2021). Inter simple sequence repeat (ISSR) markers take advantage of the regular occurrence of microsatellite repeats in the genome and have been widely used by researchers to efficiently analyze genetic diversity. This technique is a simple, fast, effective and highly repeatable method. Primers are not proprietary and can be easily designed and synthesized (Nybom 2004). Different studies have used ISSRs in plant population analysis (e.g., Luz et al. 2020; Akhtar et al. 2021; Shakoor et al. 2022) because they are more variable and need less time and money compared to other molecular markers (Harris 1999).
There are several studies evaluating genetic diversity of taxa within the genus Astragalus (Alexander et al. 2004; Rogenski et al. 2009; Anand et al. 2010; Vicente et al. 2011; Wu et al. 2019; Bagheri et al. 2020; Szabo Pamfil et al. 2021) however, A. cyclophyllon is not among these taxa. Here, morphological and genetic characteristics of A. cyclophyllon were analyzed to understand the species’ diversity. This should provide insights in population structure and infer the amount of genetic diversity present within the species, which may help to understand whether the species is threatened and deserves conservation measures. A representative sample of populations of A. cyclophyllon in Iran were studied for this purpose, using morphology and ISSR markers.
Materials and methods
Taxon sampling
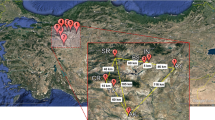
Eighty individuals (A1–A80) belonging to 29 populations (P1–P29) of A. cyclophyllon (Fig. 1) were collected from Qazvin, Hamadan, Kurdistan, and Isfahan provinces during 2019 and 2020 (Supplementary Information, Table S1) and included in this study. Leaves were silica dried in the field for later DNA extraction. Specimens of all individuals were used for morphological analyses and afterwards deposited in the herbarium of the University of Isfahan (HUI).
Morphological analyses
For each individual, we evaluated 33 morphological characters (Supplementary Information, Table S2). Among them, standard petal shape, bract length, bracteole color, indumentum of ovary and style (with/without hairs) were diagnostic features. Morphological measurements were tested for significant differences with one-way ANOVA (Kruskal–Wallis) and pairwise Mann–Whitney U test. To see if morphology can be used to group individuals within A. cyclophyllon, a two-step parsimony analysis (Bagheri et al. 2023) was conducted using the heuristic search algorithm in Paup* 4.0a169 (Swofford 2002) with initially 1000 random addition sequences collecting not more than 20 trees for each replicate. The obtained 280 trees were then used as starting trees in a heuristic search restricting the sampled tree number to 20,000.
DNA extraction and PCR amplifications
Genomic DNA was extracted from leaf tissue according to Gawel and Jarret (1991). For performing ISSR analysis, 12 ISSR primers (Blair et al. 1999) were tested, from which eight (plus two primer combinations) with reliably detectable and polymorphic DNA fragments were selected (Table 1). The PCR amplification for ISSR was performed in 15 µL volume with 3.5 µl dH2O, 7.5 µl 2 × Master Mix Red (Ampliqon) containing Taq polymerase, 2 µl primer (100 pM/mL) and 2 µl of genomic DNA (50–100 ng). After 2.5 min at 94 °C, PCR was followed by 40 cycles of 1 min at 94 °C, 1 min at the primer-specific annealing temperature (see Table 1), 5–7 min at 72 °C, and a final extension step of 7 min at 72 °C. PCR products were screened on 2% agarose gels stained with ethidium bromide. A 100 bp ladder (Thermo) was included as size reference. After running, the gels were UV visualized and recorded using a gel documentation system (GeneFlash).
Analysis of ISSR fragments
Absence and presence of clear ISSR bands in the gel was scored as “0” and “1”, respectively. A neighbor-joining (NJ) tree was calculated in Paup* 4.0a169 (Swofford 2002) using Nei-Li distances for fragments, i.e. only shared presence of bands was taken into account. Support values for the clusters we obtained through 1000 bootstrap re-samples with the same settings as before. Basic population genetic parameters including Nei’s gene diversity index (h), Shannon index (I), the percentage of polymorphic loci (PPL), and genetic differentiation indices were calculated using Popgene (version 1.32; Yeh et al. 1999). Polymorphic information content (PIC) for each primer was obtained through PowerMarker (version 3.25; Liu and Muse 2005) using the formula PIC = 1 − ∑pi2 where pi is the frequency of the ith allele (Powell et al. 1996). The correlation between genetic distances and geographic distances (r) was measured using Mantel test statistics (Mantel 1967) with 999 permutations implemented in GenAlEx version 6.5 (Peakall and Smouse 2006). An analysis of molecular variance (AMOVA) was conducted to measure the extent of intra-population and inter-population genetic diversity using GenAlEx. Additionally, a principal coordinate analysis (PCoA) was performed in GenAlEx. Population differentiation and gene flow (Nm) were estimated through FST and GST using GenAlEx. To survey the genetic population structure, Bayesian population assignment analysis was performed using Structure version 2.3 (Pritchard et al. 2000) at the population level. This method uses a Markov Chain Monte Carlo (MCMC) calculation to group individuals into populations depending on multi-locus genotype data (Falush et al. 2003). We run Structure for values of K = 2 to K = 8. As best value we obtained K = 3 with Structure Harvester (Earl and VonHoldt 2012) using the method of Evanno et al. (2005).
Results
Genetic diversity of 80 A. cyclophyllon individuals were successfully assessed using eight ISSR primers and two primer combinations. We scored 240 clear bands which all were polymorphic. The size of these bands ranged from 100 to 3000 bp. The number of polymorphic bands varied from 19 to 31 bands per primer among all populations. Primer ISSR 808, with 31 bands, produced the highest and primers ISSR 6 and UBC 873 with 19 bands each the lowest number of fragments. The average number of bands per primer for each sample was 24 (Table 2).
By calculating the ratio of the number of effective alleles to the number of observed alleles (Ne/Na), the uniformity of each population can be obtained. In this study, this ratio ranged from 0.89 to 0.97. It can be concluded that the alleles are relatively evenly distributed among A. cyclophyllon populations.
The pairwise genetic diversity using the Nei index (h) is between 0.0437 and 0.158. The average of gene diversity (h) was 0.10. The Shannon index (I) is observed between 0.0638 and 0.232 and the average of the Shannon index was 0.15. Populations P8 (Khosro Abad, Hamadan) and P25 (Cheshmeh Langan, Isfahan) showed the highest and lowest genetic diversity, respectively (Table 3). The measures for population differentiation FST and GST resulted 0.348 and 0.516, respectively, and gene flow (Nm) 0.468, indicating clear separation of populations.
The analyzed individuals of A. cyclophyllon were assigned to three genetic clusters (K = 3) by Bayesian population assignment analysis in Strucutre (Fig. 2). Populations from Hamadan, Kurdistan, and Qazvin were unified in the green group. Most populations from Isfahan province belong to a single (red) group, while populations from Hamadan, Kurdistan, plus some Isfahan individuals were grouped together (blue). Some individuals showed a signal of introgression (Fig. 2).
A Bayesian population assignment analysis based on the ISSR markers groups the 80 individuals out of 29 populations of A. cyclophyllon in three units (K = 3). B Map with the populations occurrence sites for A. cyclophyllon. The color of the population in the map corresponds to the main genetic clusters in the Structure bar plot above
Cluster analysis of 80 individuals out of 29 populations was performed based on pairwise Nei-Li distances and NJ clustering using Paup*. In the tree (Fig. 3), the samples were divided into four clusters. Cluster I contains individuals collected in populations in Hamadan and Kurdistan. Cluster II consists of the individuals from Qazvin plus one from Hamadan (A18).
Unrooted dendrogram derived from a neighbor-joining cluster analysis of 80 individuals of Astragalus cyclophyllon using ISSR markers. The horizontal bars indicate population assignment of individuals according to a Structure analysis with K = 3. Geographic origins of the individuals for four regions of Iran are provided by the vertical bars. Bootstrap support values for all groups are 100% throughout the tree
The members of both clusters are all grouped in one population (green) in the Structure analysis, although two individuals of cluster I and nearly all individuals of cluster II showed signals of introgression. Cluster III comprises only individuals from the Isfahan area in Central Iran. This cluster also belongs to a single population (red) according to Structure. Cluster IV harbors individuals from northwestern Iran, i.e. Hamadan and Kurdistan populations, together with individuals from Isfahan. Again, in the Structure analysis they are grouped in a single population (blue).
The Mantel test showed a positive correlation between genetic distance and geographic distance (r = 0.288, p = 0.02). The results of molecular analysis of variance showed that 25% of the differences were related to inter-population diversity and 75% were related to intra-population differences (Table 4).
The percentage of variation explained in PCoA for the 29 populations is 27.18% and 22.51% for the first and second axes, respectively. In the PCoA plot (Fig. 4), the samples were divided into two or three (weak) groups. According to the PCoA diagram, the populations of Okhtachi (P6) and Khosro Abad (P8) are genetically quite distant from each other and Salavat Abad A (P11) and Salavat Abad B (P12) populations are very similar. The results of this analysis are consistent with the results of the distance matrix based on the Nei index (highest genetic similarity = 0.95 between P11 and P12; lowest genetic similarity = 0.74 between P6 and P8).
Our analysis of 33 morphological traits was not able to detect clear groups within A. cyclophyllon. A maximum parsimony analysis (Supplementary Information, Fig. S1) could resolve some similar individuals from within few populations. However, most of the individuals group together in a large unresolved polytomy indicating high homoplasy for the scored characters among populations, which are anyway to a large extent quantitative.
Discussion
We investigated the genetic diversity of 80 individuals from 29 representative populations of A. cyclophyllon using molecular ISSR markers. ISSR primers amplified fragments ranging in size from 100 to 3000 bp. The total number of scored bands was 240 bands, which all were polymorphic. According to the results, the highest observed similarity coefficient (0.95) belonged to two populations (P11 and P12) from the Salavat Abad pass in the Kurdistan province and the lowest similarity was between populations (P6) Okhtachi and (P8) Khosro Abad from Hamadan (0.74). The high genetic similarity indicates ongoing gene flow between these populations or going back to a common stock of founder individuals. This is consistent with the two populations being only a limited geographical distance from each other away and occurring under very similar environmental conditions. Generally, genetic distances between the studied populations are mostly proportional to geographical distance. The results of molecular analysis of variance showed that 75% of the genetic diversity identified was within populations and 25% was between populations. Thus, genetic differentiation between the populations is relatively high (GST = 0.516). This reflects the topographic situation of the regions, which together with the distribution of the species over large geographical distances result in low amounts of gene flow (Shi et al. 2008) and nearly independent evolution of such populations.
There are different studies on genetic diversity of Astragalus species by different DNA-based molecular markers (ISSR, SRAP, SSR, RAPD). In most of them, ISSR markers could distinguish the relationships among close populations (Rogenski et al. 2009; Anand et al. 2010; Bagheri et al. 2020; Vicente et al. 2011). This is also the case for our study here, where ISSRs inferred close relationships among geographically close populations and separation among populations from different geographic areas. Some of the molecular studies on Astragalus indicated high amount of gene flow among populations (Alexander et al. 2004; Vicente et al. 2011; Szabo Pamfil et al. 2021). In A. cyclophyllon we see only restricted gene flow among populations, most probably due to the large distances between populations and specific geographic conditions of Iran such as geographical barriers (Alborz and Zagros Mountains). A similar situation was found in the threatened American Astragalus crassicarpus Nutt. in Illinois, where genetic differentiation among populations was also high when they were separated by geographical barriers (Rogenski et al. 2009).
According to the results of Structure analysis, two populations from Hamadan, five populations from Kurdistan, and three populations from Isfahan were assigned to one genetic group (blue), three other populations from Kurdistan together with seven populations from Hamadan form a second group (green). This might indicate a former geographic separation within these areas in northwestern Iran resulting in both genotype groups, followed by population expansion and a reciprocal colonization afterwards. Particularly individuals of the Qazvin populations show signals of introgression from the blue Structure group. Taking Structure results together with the cluster analysis (Fig. 3), the occurrence of blue genotypes in the more remote Isfahan area (Fig. 2) would then be due to long-distance dispersal. Nine other populations from the Isfahan area were all assigned to a clearly separate group (red), similar to the results of cluster analysis and PCoA. Some of these individuals show introgression by the blue Structure genotype, which most probably indicate introduction of the blue genotype into the area that was already populated by the red populations, where hybridization between them occurred. We assume that recent trade of animals among herders from different regions could contribute to zoochory, resulting in long-distance dispersal of A. cyclophyllon seeds.
Although we found three genotype groups by Structure analysis, which are similarly represented in the cluster analysis, separation of these groups is not accompanied by morphological differences. It seems that plasticity of morphological characters is high, probably influenced also by the differences in local habitat conditions. Moreover, although genetic differentiation is high in A. cyclophyllon, gene flow seems to occur at a low level. This should prevent differentiation of the populations from different regions of the distribution area of the species and contributes to a larger effective population size within A. cyclophyllon.
Conclusions
According to our results, A. cyclophyllon populations are genetically rather diverse and show no indication of inbreeding depression. Long-distance dispersal and gene flow among northern and southern populations was detected (Fig. 2), which keeps populations united and results in an effectively large gene pool. The most important threat for this species seems therefore the high grazing pressure on the habitats of the taxon. This could result in reduced establishment of seedlings and, over time, an alteration of population demography towards ageing stands with only very few young individuals. Thus, management strategies for conservation of A. cyclophyllon should be directed to the herders to educate them about a sustainable stock size of livestock that a certain area can carry.
Data availability
All data generated or analyzed during this study are included in the paper or supplementary information.
References
Akhtar N, Hafiz IA, Hayat MQ, Potter D, Abbasi NA, Habib U, Hussain A, Hafeez H, Bashir MA, Malik SI (2021) ISSR-based genetic diversity assessment of genus Jasminum L. (Oleaceae) from Pakistan. Plants 10:1270
Alexander JA, Liston A, Popovich SJ (2004) Genetic diversity of the narrow endemic Astragalus oniciformis (Fabaceae). Am J Bot 91:2004–2012
Anand KK, Sivastava RK, Chaudhary LB, Singh AK (2010) Delimitation of species of the Astragalus rhizanthus complex (Fabaceae) using molecular markers RAPD, ISSR, and DAMD. Taiwania 55:197–207
Bagheri A, Abbasi S, Mahmoodi M, Akhavan Roofigar A, Blattner FR (2020) Genetic structure and conservation status of Astragalus subrecognitus (Fabaceae): a very rare and narrow endemic species. Plant Ecol Evol 153:101–107
Bagheri A, Maassoumi AA, Brassac J, Blattner FR (2023) Dated phylogeny of Astragalus section Stereothrix (Fabaceae) and allied taxa in the Hypoglottis clade. Biology 12:138
Blair MW, Panaud O, McCouch SR (1999) Inter simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor Appl Genet 9:780–792
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Gawel NJ, Jarret RL (1991) A modified CTAB extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep 9:262–266
Harris J (1999) RAPDs in systematics—a useful methodology? In: Hollingsworth PM, Bateman RM, Gornall RJ (eds) Molecular systematics and plant evolution. Systematics Association, London, pp 211–228
Hasan N, Choudhary S, Naaz N, Sharma N, Laskar RA (2021) Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J Genet Eng Biotechnol 19:128
Jalili A, Jamzad Z (1999) Red data book of Iran, a preliminary survey of endemic, rare and endangered plant species in Iran. Research Institute of Forest and Rangelands Press, Tehran
Kalpana J, Warude PC, Bhushan P (2004) Molecular markers in herbal drug technology. Curr Sci 87:159–165
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Lombard V, Dubreuil P, Dillman C, Baril C (2001) Genetic distance estimators based on molecular data for plant registration and protection: a review. Acta Hortic 546:55–63
Luz GC, Strioto DK, Mangolin CA, Machado MDFPS (2020) ISSR markers to assess genetic diversity of cultivated populations from artificial selection of Stevia rebaudiana (Bert.) Bertoni. Breed Sci 70:508–514
Maassoumi AA (1998) Astragalus in the old world: check-list. Research Institute of Forests and Rangelands, Tehran
Maassoumi AA (2005) The genus Astragalus in Iran, vol 5. Research Institute of Forests and Rangelands, Tehran
Maassoumi AA (2020) http://astragalusofworld.com. Accessed 10 August 2022
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Podlech D (1986) Taxonomic and phytogeographical problems in Astragalus of the Old World and South-West Asia. Proc Royal Soc B 89:37–43
Podlech D, Zarre S (2013) Taxonomic revision of the genus Astragalus L. (Leguminosae) in the old world, vol 2. Naturhistorisches Museum Wien, Vienna
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rogenski C, Smith M, Esselman E (2009) ISSR marker diversity of the threatened Astragalus crassicarpus var. trichocalyx in Illinois populations. Trans Ill State Acad Sci 102:149–159
Shakoor A, Zaib G, Zhao F, Li W, Lan X, Esfandani-Bozchaloyi S (2022) ISSR markers and morphometry determine genetic diversity and population structure in Hedera helix L. Czech J Genet Plant Breed 58:73–82
Shi W, Yang CF, Chen JM, Guo YH (2008) Genetic variation among wild and cultivated populations of the Chinese medicinal plant Coptis chinensis (Ranunculaceae). Plant Biol 10:485–491
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland
Szabo Pamfil D, Badarau AS, Harta M (2021) Assessment of genetic diversity and population structure of the endangered Astragalus exscapus subsp. transsilvanicus through DNA-based molecular markers. Plants 10:2732
Vicente MJ, Segura F, Aguado M, Migliaro D, Franco JA, Martínez-Sánchez JJ (2011) Genetic diversity of Astragalus nitidiflorus, a critically endangered endemic of SE Spain, and implications for its conservation. Biochem Syst Ecol 39:175–182
Wu M, Shi X, Tan D (2019) Development of SSR markers for Astragalus lehmannianus, a vulnerable species from northwestern China. Appl Plant Sci 7:1–4
Yeh F, Boyle T, Rongcai Y, Ye Z (1999) POPGENE: Microsoft window-based freeware for population genetic analysis version 1.3. University of Alberta Press, Edmonton
Acknowledgements
We wish to thank the University of Isfahan for supporting this study.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aliabadi, F., Bagheri, A., Abbasi, S. et al. High genetic diversity in an endemic and vulnerable species: evidence from Astragalus cyclophyllon (Fabaceae) in Iran. Genet Resour Crop Evol 70, 1999–2008 (2023). https://doi.org/10.1007/s10722-023-01550-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01550-7