Abstract
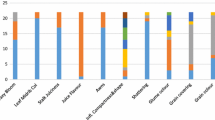
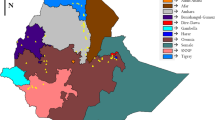
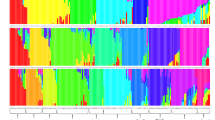
Ethiopia is considered to be the center of origin and diversity for sorghum [Sorghum bicolor (L.) Moench]. Knowledge of the genetic structure of the crop is important for its improvement and proper conservation. Therefore, the present study targeted to explore the genetic diversity and population structure of 92 Ethiopian sorghum genotypes representing five populations using 12 polymorphic microsatellite markers. The study resulted in a total of 77 alleles across the entire loci and populations. All the used microsatellite loci were highly polymorphic with PIC ranging from 0.66 to 0.82 with overall mean of 0.76. The analysis confirmed the presence of high within-population diversity with Nei’s gene diversity ranging from 0.71 to 0.84 with overall mean of 0.79. Analysis of molecular variance (AMOVA) confirmed high genetic differentiation (FST = 0.29) where 90% of the total genetic variation resides within populations, leaving only 10% among populations. The PCoA, clustering, and population structure did not cluster the studied populations into sharply genetically distinct clusters according to their geographical areas of sampling due to the presence of considerable gene flow (Nm = 2.13). Overall, the used microsatellite loci were highly informative and hence, a useful genetic tool to investigate the genetic structure of sorghum. Among the five studied, populations from North Gondar (Het = 0.75) and South Tigray region (Het = 0.74) showed the highest genetic diversity, and hence these areas could be considered as hot spots for identification of supper-performing genotypes to be used in sorghum breeding program, and also for designing appropriate germplasm conservation strategies.






Similar content being viewed by others
Data availability
All data generated or analyzed as part of this study are included in this published article.
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- FIS:
-
Inbreeding coefficient
- FIT:
-
Overall inbreeding coefficient
- FST:
-
Fixation index
- GD:
-
Gene diversity
- He:
-
Heterozygosity
- I:
-
Shannon’s index
- MAF:
-
Major allele frequency
- Na:
-
Number of allele
- NABRC:
-
National agricultural biotechnology research center
- Ne:
-
Number of effective alleles
- Nm:
-
Gene flow
- PAGE:
-
Polyacrylamide gel electrophoresis
- PCoA:
-
Principal cordinate analysis
- PIC:
-
Polymorphic information content
- SSR:
-
Simple sequence repeats
References
Adugna A (2014) Analysis of in situ diversity and population structure in Ethiopian cultivated Sorghum bicolor (L.) landraces using phenotypic traits and SSR markers. Springerplus 3(1):1–14
Adugna A, Snow AA, Sweeney PM, Bekele E, Mutegi E (2013) Population genetic structure of in situ wild Sorghum bicolor in its Ethiopian center of origin based on SSR markers. Genet Resour Crop Evol 60(4):1313–1328
Afolayan G, Deshpande SP, Aladele SE, Kolawole AO, Angarawai I, Nwosu DJ, Michael C, Blay ET, Danquah EY (2019) Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) accessions using single nucleotide polymorphism markers. Plant Genetic Resour 17(5):412–420
Ali ML, Rajewski JF, Baenziger PS, Gill KS, Eskridge KM, Dweikat I (2008) Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Mol Breeding 21(4):497–509
Almekinders CJM, Elings A (2001) Collaboration of farmers and breeders: Participatory crop improvement in perspective. Euphytica 122(3):425–438
Appa Rao S, Prasada Rao KE, Mengesha MH, Reddy VG (1996) Morphological diversity in sorghum germplasm from India. Genet Resour Crop Evol 43(6):559–567
Ayana A, Bryngelsson T, Bekele E (2000) Genetic variation of Ethiopian and Eritrean sorghum (Sorghum bicolor (L.) Moench) germplasm assessed by random amplified polymorphic DNA (RAPD). Genetic Resour Crop Evol 47(5):471–482
Billot C, Ramu P, Bouchet S, Chantereau J, Deu M, Gardes L, Noyer JL, Rami JF, Rivallan R, Li Y, Lu P (2013) Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLoS ONE 8(4):e59714
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314
Bucheyeki TL, Gwanama C, Mgonja M, Chisi M, Folkertsma R, Mutegi R (2009) Genetic variability characterisation of Tanzania sorghum landraces based on simple sequence repeats (SSRs) molecular and morphological markers. Afr Crop Sci J. https://doi.org/10.4314/acsj.v17i2.54201
Burow G, Franks CD, Xin Z, Burke JJ (2012) Genetic diversity in a collection of Chinese sorghum landraces assessed by microsatellites. Am J Plant Sci. https://doi.org/10.4236/ajps.2012.312210
Cuevas HE, Prom LK (2020) Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan Sorghum Core collection. BMC Genomics 21(1):1–15
Cuevas HE, Prom LK, Cooper EA, Knoll JE, Ni X (2018) Genome-wide association mapping of anthracnose (Colletotrichum sublineolum) resistance in the US sorghum association panel. Plant Genome 11(2):170099
Disasa T, Feyissa T, Admassu B (2017) Characterization of Ethiopian sweet sorghum accessions for 0brix, morphological and grain yield traits. Sugar Tech 19(1):72–82
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
El-Amin HKA, Hamza NB (2013) Phylogenetic diversity of Sorghum bicolor (L.) Moench accessions from different regions in Sudan. Am J Biochem Mol Biol 3(1):127–134
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Evans LT (1984) Darwin’s use of the analogy between artificial and natural selection. J Hist Biol 17:113–140
FAOSTAT (2019). Food and agriculture organization of the United Nations. Database. Available at: http://faostat.fao.org/databas
Gasura E, Setimela PS, Souta CM (2015) Evaluation of the performance of sorghum genotypes using GGE biplot. Can J Plant Sci 95(6):1205–1214
Ghebru B, Schmidt R, Bennetzen J (2002) Genetic diversity of Eritrean sorghum landraces assessed with simple sequence repeat (SSR) markers. Theor Appl Genet 105(2):229–236
Girma A, Seo W, She RC (2019) Antibacterial activity of varying UMF-graded Manuka honeys. PLoS ONE 14(10):e0224495
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genetics Res Int. https://doi.org/10.1155/2015/431487
Greenbaum G, Templeton AR, Zarmi Y, Bar-David S (2014) Allelic richness following population founding events–a stochastic modeling framework incorporating gene flow and genetic drift. PLoS ONE 9(12):e115203
Huang BE, Verbyla KL, Verbyla AP, Raghavan C, Singh VK, Gaur P, Leung H, Varshney RK, Cavanagh CR (2015) MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128(6):999–1017
Jordan DR, Tao YZ, Godwin ID, Henzell RG, Cooper M, McIntyre CL (1998) Loss of genetic diversity associated with selection for resistance to sorghum midge in Australian sorghum. Euphytica 102(1):1–7
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Kristensen PS, Jahoor A, Andersen JR, Cericola F, Orabi J, Janss LL, Jensen J (2018) Genome-wide association studies and comparison of models and cross-validation strategies for genomic prediction of quality traits in advanced winter wheat breeding lines. Front Plant Sci 9:69
Labeyrie V, Deu M, Barnaud A, Calatayud C, Buiron M, Wambugu P, Manel S, Glaszmann JC, Leclerc C (2014) Influence of ethnolinguistic diversity on the sorghum genetic patterns in subsistence farming systems in Eastern Kenya. PLoS ONE 9(3):e92178
Leff B, Ramankutty N, Foley JA (2004) Geographic distribution of major crops across the world. Global Biogeochem Cycles. https://doi.org/10.1029/2003GB002108
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Mace ES, Rami JF, Bouchet S, Klein PE, Klein RR, Kilian A, Wenzl P, Xia L, Halloran K, Jordan DR (2009) A consensus genetic map of sorghum that integrates multiple component maps and high-throughput diversity array technology (DArT) markers. BMC Plant Biol 9(1):1–14
Manzelli M, Pileri L, Lacerenza N, Benedettelli S, Vecchio V (2007) Genetic diversity assessment in Somali sorghum (Sorghum bicolor (L.) Moench) accessions using microsatellite markers. Biodivers Conserv 16(6):1715–1730
Mathur S, Umakanth AV, Tonapi VA, Sharma R, Sharma MK (2017) Sweet sorghum as biofuel feedstock: recent advances and available resources. Biotechnol Biofuels 10(1):1–19
Mengistu MG, Steyn JM, Kunz RP, Doidge I, Hlophe HB, Everson CS, Jewitt GPW, Clulow AD (2016) A preliminary investigation of the water use efficiency of sweet sorghum for biofuel in South Africa. Water SA 42(1):152–160
Missihoun AA, Adoukonou-Sagbadja H, Sedah P, Agbangla C, Ahanhanzo C, Dagba RA (2015) Genetic diversity of Sorghum bicolor (L.) Moench landraces from Northwestern Benin as revealed by microsatellite markers. Afr J Biotechnol 14(16):1342–1353
Mofokeng A, Shimelis H, Tongoona P, Laing M (2014) A genetic diversity analysis of South African sorghum genotypes using SSR markers. South Afr J Plant Soil 31(3):145–152
Motlhaodi T, Geleta M, Chite S, Fatih M, Ortiz R, Bryngelsson T (2017) Genetic diversity in sorghum [Sorghum bicolor (L.) Moench] germplasm from Southern Africa as revealed by microsatellite markers and agro-morphological traits. Genetic Resour Crop Evol 64(3):599–610
Muui CW, Muasya RM, Kirubi DT, Runo SM, Karugu A (2016) Genetic variability of sorghum landraces from lower Eastern Kenya based on simple sequence repeats (SSRs) markers. Afr J Biotech 15(8):264–271
Ng’uni D, Geleta M, Bryngelsson T (2011) Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) accessions of Zambia as revealed by simple sequence repeats (SSR). Hereditas 148(2):52–62
Ng'uni D, Geleta M, Hofvander P, Fatih M, Bryngelsson T (2012) Comparative genetic diversity and nutritional quality variation among some important Southern African sorghum accessions ['Sorghum bicolor'(L.) Moench]. Aust J Crop Sci, 6(1):56–64
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in excel population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Perrier X, Flori A (2003) Methods of data analysis. In: Press CRC (ed) Genetic diversity of cultivated tropical plants. pp 47–80
Poehlman JM, Sleper DA, (1979) Breeding field crops. eds (No. 631.5220 POE 1995. CIMMYT)
Pritchard JK, Wen W, Falush D (2003) Documentation for the structure software, version 2. Department of Human Genetics, University of Chicago, Chicago
Ramu P, Billot C, Rami JF, Senthilvel S, Upadhyaya HD, Ananda Reddy L, Hash CT (2013) Assessment of genetic diversity in the sorghum reference set using EST-SSR markers. Theor Appl Genet 126(8):2051–2064
Reddy BV, Ramesh S, Reddy PS, Kumar AA, Tropics SA (2009) Genetic enhancement for drought tolerance in sorghum. Plant Breed Rev 31:189–222
Rhodes DH, Hoffmann L, Rooney WL, Herald TJ, Bean S, Boyles R, Brenton ZW, Kresovich S (2017) Genetic architecture of kernel composition in global sorghum germplasm. BMC Genomics 18(1):1–8
Ritter KB, McIntyre CL, Godwin ID, Jordan DR, Chapman SC (2007) An assessment of the genetic relationship between sweet and grain sorghums, within Sorghum bicolor ssp. bicolor (L.) Moench, using AFLP markers. Euphytica 157(1):161–176
Semahegn Z (2019) The genetic improvement of sorghum in Ethiopia: review. JBAH [Internet]. 2019 Feb [cited 2022 Mar 3]
Shete S, Tiwari H, Elston RC (2000) On estimating the heterozygosity and polymorphism information content value. Theor Popul Biol 57(3):265–271
Singh SP (1985) Sources of cold tolerance in grain sorghum. Can J Plant Sci 65(2):251–257
Singh R, Axtell JD (1973) High lysine mutant gene (hl that improves protein quality and biological value of grain sorghum 1. Crop Sci 13(5):535–539
Statista (2020). Sorghum production worldwide in 2019/2020, by leading country (in 1,000 metric tons). Hamburg: Statista
Swarup S, Cargill EJ, Crosby K, Flagel L, Kniskern J, Glenn KC (2021) Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci 61(2):839–852. https://doi.org/10.1002/csc2.20377
Tesfamichael TA, Githiri SM, Kasili RW, Skilton RA, Solomon M, Nyende AB (2014) Genetic diversity analysis of Eritrean sorghum (Sorghum bicolor (L.) Moench) germplasm using SSR markers. Mol Plant Breed. https://doi.org/10.5376/mpb.2014.05.0013
Thomas M, Demeulenaere E, Dawson JC, Khan AR, Galic N, Jouanne-Pin S, Remoué C, Bonneuil C, Goldringer I (2012) On-farm dynamic management of genetic diversity: the impact of seed diffusions and seed saving practices on a population-variety of bread wheat. EvOl Appl 5(8):779–795
Upadhyaya HD, Reddy KN, Vetriventhan M, Ahmed MI, Krishna GM, Reddy MT, Singh SK (2017) Sorghum germplasm from West and Central Africa maintained in the ICRISAT genebank: status, gaps, and diversity. Crop J 5(6):518–532
Vus NA, Kobyzeva LN, Bezuglaya ON (2020) Determination of the breeding value of collection chickpea (Cicer arietinum L) accessions by cluster analysis. Vavilov J Genet Breed 24(3):244
Wang J, Duncan D, Shi Z, Zhang B (2013) WEB-based gene set analysis toolkit (WebGestalt): update 2013. Nucleic acids research, 41(W1):W77-W83
Wang ML, Zhu C, Barkley NA, Chen Z, Erpelding JE, Murray SC, Tuinstra MR, Tesso T, Pederson GA, Yu J (2009) Genetic diversity and population structure analysis of accessions in the US historic sweet sorghum collection. Theor Appl Genet 120(1):13–23
Weerasooriya DK, Maulana FR, Bandara AY, Tirfessa A, Ayana A, Mengistu G, Nouh K, Tesso TT (2016) Genetic diversity and population structure among sorghum (Sorghum bicolor, L.) germplasm collections from Western Ethiopia. Afr J Biotechnol 15(23):1147–1158
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgements
We would like to thank the Institute of Biotechnology of Addis Ababa University and the National Agricultural Biotechnology Research Center of the Ethiopian Institute of Agricultural Research (EIAR) for the complete financial support and for providing laboratory facilities to carry out this research.
Funding
This study was funded by the integrated research grants of EIAR-NABRC (Ethiopian Institute of Agricultural Research-National Agricultural Biotechnology Research Center) and Addis Ababa University of the Ethiopian Minister of Science and Higher Education.
Author information
Authors and Affiliations
Contributions
AM has contributed to the study material collection and generation of row data under laboratory conditions. All authors have equally participated in the design of the study, data analysis, and interpretation, drafting of the manuscript, and revising and commenting on the manuscript critically for final submission.
Corresponding authors
Ethics declarations
Conflict of interest
The authors disclose that they do not have any competing interests.
Ethical approval
We check that all named authors have read and approved the paper and that no other individuals who meet the criteria for authorship but are not listed have done so. We also affirm that the manuscript's authorship order has been approved by all of us.
Ethical responsibility
The data in this manuscript is unique to us and has never been published before. Other people's work and words have been acknowledged appropriately.
Human or animal rights
There are no human or animal participants in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Misganaw, A., Feyissa, T., Mekonnen, T. et al. Genetic diversity analysis of sorghum genotypes for sustainable genetic resource conservation and its implication for breeding program in ethiopia. Genet Resour Crop Evol 70, 1831–1852 (2023). https://doi.org/10.1007/s10722-023-01539-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01539-2