Abstract
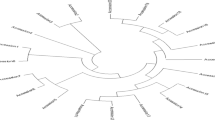
Sweet sorghum (Sorghum bicolor L.) is a type of cultivated sorghums and has been recognized widely as potential alternative source of bio-fuel because of its high fermentable sugar content in the stalk. A substantial variation of sugar content and related traits is known to exist in US sweet sorghum. The objectives of the study were to assess the genetic diversity and relationship among the US sweet sorghum cultivars and lines using SSR markers and to examine the genetic variability within sweet sorghum accessions for sugar content. Sixty-eight sweet sorghum and four grain sorghum cultivars and lines were genotyped with 41 SSR markers that generated 132 alleles with an average of 3.22 alleles per locus. Polymorphism information content (PIC) value, a measure of gene diversity, was 0.40 with a range of 0.03–0.87. The genetic similarity co-efficient was estimated based on the segregation of the 132 SSR alleles. Clustering analysis based on the genetic similarity (GS) grouped the 72 sorghum accessions into 10 distinct clusters. Grouping based on clustering analysis was in good agreement with available pedigree and genetic background information. The study has revealed the genetic relationship of cultivars with unknown parentage to those with known parentage. A number of diverse pairs of sweet sorghum accessions were identified which were polymorphic at many SSR loci and significantly different for sugar content as well. Information generated from this study can be used to select parents for hybrid development to maximize the sugar content and total biomass, and development of segregating populations to map genes controlling sugar content in sweet sorghum.

Similar content being viewed by others
References
Agrama HA, Tuinstra MR (2003) Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr J Biotechnol 2:334–340
Ahmad M. (2002) Assessment of genomic diversity among wheat genotypes as determined by simple sequence repeats. Genome 45:646–651
Alamnza-Pinzon MI, Khairullah M, Fox PN, Warburton ML (2003) Comparison of molecular markers and co-efficients of parentage for the analysis of genetic diversity among spring bread wheat accessions. Euphytica 130:77–86
Anderson JA, Churchill GA, Autrique JE, Sorrellls ME, Tanksley SD (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Becelaere GV, Edward LL, Paterson AH, Chee PW (2005) Pedigree-vs. DNA marker-based genetic similarity estimates in Cotton. Crop Sci 45:2281–2287
Bohn M, Utz HF, Melchinger AE (1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs and SSRs and their use for predicting progeny variance. Crop Sci 39:228–237
Budak H, Pedraza F, Cregan PB, Baenziger PS, Dweikat I (2003) Development and utilization of SSRs to estimate the degree of genetic relationships in a collection of pearlmillet germplasm. Crop Sci 43:2284–2290
Casa AM, Mitchell SE, Hamblin MT, Sun H, Bowers JE, Paterson AH, Aquardo CF, Kresovich S (2005) Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theor Appl Genet 111:23–30
Cox TS, Murphy JP, Rodgers DM (1986) Changes in genetic diversity in the red winter wheat regions of the United States. Proc Natl Acad Sci USA 83:5583–5586
Dean RE, Dahlberg JA, Hopkins MS, Mitchell SE, Kresovich (1999) Genetic redundancy and diversity among Orange accessions in the US national sorghum collection as assessed with simple sequence (SSR) markers. Crop Sci 39:1215–1221
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Fufa H, Baenziger PS, Beecher BS, Dweikat I, Graybosch, Eskridge KM (2005) Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145:133–146
Gnansounou E, Dauriat A, Wyman C (2005) Refining sweet sorghum to ethanol and sugar: economic trade-offs in the context of North China. Bioresoure Tech 96:985–1002
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Innan H, Terauchi R, Miyashita T (1997) Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Kuleung C, Baenziger PS, Dweikat I (2004) Transferability of SSR markers among wheat, rye and triticale. Theor Appl Genet 108:1147–1150
Kuleung C, Baenziger PS, Kachman SD, Dweikat I (2006) Evaluating the genetic diversity of triticale with wheat and rye SSR markers. Crop Sci 46:1692–1700
Mahmood A, Baeeziger PS, Budak H, Gill KS, Dweikat I (2004) The use of microsatellite markers for the detection of genetic similarity among winter bread wheat lines for chromosome 3A. Theor Appl Genet 109:1494–1503
Melchinger AE (1999) Genetic diversity and heterosis. In: The genetics and exploitation of heterosis in crops. American Society of Agronomy, Madison, Wisc., pp 99–118
Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullet JE (2004) Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci 44:1236–1244
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl acad Sci USA 76:5269–5273
Parker GD, Fox PN, Langridge P, Chalmers K, Whan B, Ganter PF (2002) Genetic diversity within Australian wheat breeding programs based on molecular and pedigree data. Euphytica 124:293–306
Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system. Exeter Publishing, Setauket
SAS Institure (1999–2001) The SAS system for windows V8. Release 8.2 (TS2MO)
Schloss SJ, Mitchell SE, White GM, Kukatla R, Bowers JE, Paterson AH, Kresovich S (2002) Characterization of RFLP probe sequences for gene discovery and SSR development in Sorghum bicolor (L.) Moench. Theor Appl Genet 105:912–920
Senior ML, Murphy JP, Goodman NM, Stuber CW (1998) Utility of SSRs for determining genetic similarities and relationships in maize using an agarose gel system. Crop Sci 38:1088–1098
Smith CW, Frederiksen RA (2000) Sorghum: origin, history, technology and production. Wiley, New York. Preface, pp vii–ix
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Smith JSC, Kresovich S, Hopkins MS, Mitchell SE, Dean RE, Woodman WL, Lee M, Porter K (2000) Genetic diversity among elite sorghum inbred lines assessed with Simple sequence repeats. Crop Sci 40:226–232
Somers DJ, Banks T, Depauw R, Clarke J, Fox S, Pozniak C (2007) Genome-wide linkage disequilibrium in wheat. Plant and Animal Genome XV conference, 13–17 Jan., 2007, San Diego, CA, Poster abstract P268
Souza E, Sorrells ME (1989) Pedigree analysis of North American Oat cultivars released from 1951 to 1985. Crop Sci 29:595–601
Tams SH, Melchinger AE, Bauer E (2005) Genetic similarity among European winter triticale elite germplasms assessed with AFLP and comparisons with SSR and pedigree data. Plant Breed 124:154–160
Van Beuningen LT, Busch RH (1997) Genetic diversity among North American spring wheat cultivars: III. Cluster analysis based on quantitative morphological traits. Crop Sci 37:981–988
Weber JL (1990) Informativeness of human (dC-dA)n (dG-dT)n poylmorphisms. Genomics 7:524–530
Weir BS (1996) Genetic data analysis: II. Methods for the discrete population genetic data, 2nd edn. Sinnauer Associates, Sunderland
Acknowledgements
This research was supported by USDA-CSREES-NRI Grant 2004-35300-14700. Authors extend special thanks to Dr. J. F. Pederson and Mr. J. J. Toy, USDA-ARS, University of Nebraska-Lincoln, Nebraska; National Center for Genetic Resources Preservation (NCGRP), 111 South Mason, Fort Collins, Colorado; National Plant Germplasm System, Griffin, Georgia; Dr. W. L. Rooney, Texas Agricultural System Station, College station, Texas and Dr. M. Bitzer, University of Kentucky, Kentucky for providing seeds of sweet sorghum cultivars and lines.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ali, M.L., Rajewski, J.F., Baenziger, P.S. et al. Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Mol Breeding 21, 497–509 (2008). https://doi.org/10.1007/s11032-007-9149-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-007-9149-z