Abstract
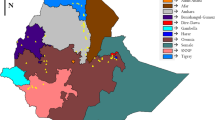
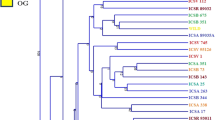
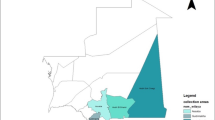
Genetic integrity of an accession should be preserved in the conservation of germplasm. Characterization of diverse germplasm based on a molecular basis enhances its conservation and use in breeding programs. The aim of this study was to assess the genetic diversity of 169 sorghum accessions using a total of 6977 SNP markers. The polymorphic information content of the markers was 0.31 which is considered to be moderately high. Structure analysis using ADMIXTURE program revealed a total of 10 subpopulations. Neighbor-joining tree revealed the presence of six main clusters among these subpopulations whereas in principal component analysis, seven clusters were identified. Cluster analysis grouped most populations depending on source of collection although other accessions originating from the same source were grouped under different clusters. Analysis of molecular variance (AMOVA) revealed 30% and 70% of the variation occurred within and among accessions, respectively. Gene flow within the populations was, however, limited indicating high differentiation within the subpopulation. Observed heterozygosity among accessions varied from 0.03 to 0.06 with a mean of 0.05 since sorghum is a self-pollinating crop. High genetic diversity among the subpopulations can be further explored for superior genes to develop new sorghum varieties.




Similar content being viewed by others
References
Abraha T, Githiri SM, Kasili RW, Skilton RA, Solomon M, Nyende AB (2014) Genetic diversity analysis of Eritrean sorghum (Sorghum bicolor (L.) Moench) germplasm using SSR markers. Mol Plant Breed 5(13):1–12. https://doi.org/10.5376/mpb.2014.05.0013
Adugna A (2014) Analysis of in situ diversity and population structure in Ethiopian cultivated Sorghum bicolor (L.) landraces using phenotypic traits and SSR markers. SpringerPlus 3(1):1–14. https://doi.org/10.1186/2193-1801-3-212
Afolayan G, Deshpande SP, Aladele SE, Kolawole AO, Angarawai I, Nwosu DJ, Michael C, Blay ET, Danquah EY (2019) Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) accessions using single nucleotide polymorphism markers. Plant Genet Resour Charact Util 17(5):412–420. https://doi.org/10.1017/s1479262119000212
Alexander DH, Lange K (2011) Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform 12(1):246. https://doi.org/10.1186/1471-2105-12-246
Ayana A, Bekele E, Bryngelsson T (2000) Genetic variation in wild sorghum (Sorghum bicolor ssp. verticilliflorum (L.) Moench) germplasm from Ethiopia assessed by random amplified polymorphic DNA (RAPD). Hereditas 132(3):249–254. https://doi.org/10.1111/j.1601-5223.2000.t01-1-00249.x
Batley J, Edwards D (2007) SNP applications in plants. In: Oraguzie NC, Rikkerink EHA, Gardiner SE, De Silva HN (eds) Association mapping in plants. Springer, New York, pp 95–102. https://doi.org/10.1007/978-0-387-36011-9_6
Blum A (2011) Plant breeding for water-limited environments. Springer, New York. https://doi.org/10.1007/978-1-4419-7491-4
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Bucheyeki T, Gwanama C, Mgonja M, Chisi M, Folkertsma R, Mutegi R (2009) Genetic variability characterisation of Tanzania sorghum landraces based on simple sequence repeats (SSRs) molecular and morphological markers. Afr Crop Sci J. https://doi.org/10.4314/acsj.v17I2.54201
Muui WM, Reuben MM, Duncan TK, Steven MR, Arthur K (2016) Genetic variability of sorghum landraces from lower Eastern Kenya based on simple sequence repeats (SSRs) markers. Afr J Biotechnol 15(8):264–271. https://doi.org/10.5897/ajb2015.14680
Clarke WE, Parkin IA, Gajardo HA, Gerhardt DJ, Higgins E, Sidebottom C, Sharpe AG, Snowdon RJ, Federico ML, Iniguez-Luy FL (2013) Genomic DNA enrichment using sequence capture microarrays: a novel approach to discover sequence nucleotide polymorphisms (SNP) in Brassica napus L. PLoS ONE. https://doi.org/10.1371/journal.pone.0081992
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. https://doi.org/10.1371/journal.pone.0019379
Eltaher S, Sallam A, Belamkar V, Emara HA, Nower AA, Salem KFM, Poland J, Baenziger PS (2018) Genetic diversity and population structure of F3:6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front Genet. https://doi.org/10.3389/fgene.2018.00076
Enyew M, Feyissa T, Carlsson AS, Tesfaye K, Hammenhag C, Geleta M (2022) Genetic diversity and population structure of sorghum (Sorghum bicolor (L.) Moench) accessions as revealed by single nucleotide polymorphism markers. Front Plant Sci. https://doi.org/10.3389/fpls.2021.799482/full
FAOSTAT (2019) FAOSTAT. https://www.fao.org/faostat/en/#data/QCL/visualize
Ferrão LFV, Caixeta ET, de F. Souza F, Zambolim EM, Cruz CD, Zambolim L, Sakiyama NS (2013) Comparative study of different molecular markers for classifying and establishing genetic relationships in Coffea canephora. Plant Syst Evol 299(1):225–238. https://doi.org/10.1007/s00606-012-0717-2
Ferrão LFV, Caixeta ET, Cruz CD, de F. Souza F, Ferrão MAG, Maciel-Zambolim E, Zambolim L, Sakiyama NS (2014) The effects of encoding data in diversity studies and the applicability of the weighting index approach for data analysis from different molecular markers. Plant Syst Evol 300(7):1649–1661. https://doi.org/10.1007/s00606-014-0990-3
Franco J, Crossa J, Ribaut JM, Bertran J, Warburton ML, Khairallah M (2001) A method for combining molecular markers and phenotypic attributes for classifying plant genotypes. Theor Appl Genet 103(6):944–952. https://doi.org/10.1007/s001220100641
Geleta N, Labuschagne MT (2005) Qualitative traits variation in sorghum (Sorghum bicolor (L.) Moench) germplasm from eastern highlands of Ethiopia. Biodivers Conserv 14(13):3055–3064. https://doi.org/10.1007/S10531-004-0315-x
Girma G, Nida H, Seyoum A, Mekonen M, Nega A, Lule D, Dessalegn K, Bekele A, Gebreyohannes A, Adeyanju A, Tirfessa A, Ayana G, Taddese T, Mekbib F, Belete K, Tesso T, Ejeta G, Mengiste T (2019) A large-scale genome-wide association analyses of Ethiopian sorghum landrace collection reveal loci associated with important traits. Front Plant Sci 10:691. https://doi.org/10.3389/fpls.2019.00691
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartr: an r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour 18(3):691–699. https://doi.org/10.1111/1755-0998.12745
Guo C, McDowell IC, Nodzenski M, Scholtens DM, Allen AS, Lowe WL, Reddy TE (2017) Transversions have larger regulatory effects than transitions. BMC Genomics 18(1):1. https://doi.org/10.1186/s12864-017-3785-4
Hamrick JL (1983) The distribution of genetic variation within and among natural plant populations. Genet Conserv 335–348
He J, Zhao X, Laroche A, Lu ZX, Liu HK, Li Z (2014) Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00484/full
Kamvar ZN, Tabima JF, Gr̈unwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014(1):1–14. https://doi.org/10.7717/peerj.281/table-6
Kumar S, Banks TW, Cloutier S (2012) SNP discovery through next-generation sequencing and its applications. Int J Plant Genomics. https://doi.org/10.1155/2012/831460
Kumar D, Chhokar V, Sheoran S, Singh R, Sharma P, Jaiswal S, Iquebal MA, Jaiswar A, Jaisri J, Angadi UB, Rai A, Singh GP, Kumar D, Tiwari R (2020) Characterization of genetic diversity and population structure in wheat using array based SNP markers. Mol Biol Rep 47(1):293–306. https://doi.org/10.1007/s11033-019-05132-8
Lasky JR, Upadhyaya HD, Ramu P, Deshpande S, Hash CT, Bonnette J, Juenger TE, Hyma K, Acharya C, Mitchell SE, Buckler ES, Brenton Z, Kresovich S, Morris GP (2015) Genome-environment associations in sorghum landraces predict adaptive traits. Sci Adv. https://doi.org/10.1126/sciadv.1400218
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296. https://doi.org/10.1093/NAR/GKAB301
Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL (2008) Prioritizing climate change adaptation needs for food security in 2030. Science 319(5863):607–610. https://doi.org/10.1126/science.1152339
Luo Z, Iaffaldano BJ, Zhuang X, Fresnedo-Ramírez J, Cornish K (2017) Analysis of the first Taraxacum kok-saghyz transcriptome reveals potential rubber yield related SNPs. Sci Rep. https://doi.org/10.1038/S41598-017-09034-2
Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ, Roth A, Bagley MJ, Nacci DE (2010) Population genetic diversity and fitness in multiple environments. BMC Evol Biol. https://doi.org/10.1186/1471-2148-10-205
Mehmood S, Bashir A, Ahmad A, Akram Z, Jabeen N, Gulfraz M (2008) Molecular characterization of regional Sorghum bicolor varieties from Pakistan. Pak J Bot 40(5):2015–2021.
Menamo T, Kassahun B, Borrell AK, Jordan DR, Tao Y, Hunt C, Mace E (2021) Genetic diversity of Ethiopian sorghum reveals signatures of climatic adaptation. Theor Appl Genet 134(2):731–742. https://doi.org/10.1007/s00122-020-03727-5
Mengistu G, Shimelis H, Laing M, Lule D, Assefa E, Mathew I (2020) Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) landraces using SNP markers. S Afr J Plant Soil 37(3):220–226. https://doi.org/10.1080/02571862.2020.1736346
Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullet JE (2004) Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci 44(4):1236–1244. https://doi.org/10.2135/cropsci2004.1236
Morton BR, Bi IV, McMullen MD, Gaut BS (2006) Variation in mutation dynamics across the maize genome as a function of regional and flanking base composition. Genetics 172(1):569–577. https://doi.org/10.1534/genetics.105.049916
Motlhaodi T, Geleta M, Chite S, Fatih M, Ortiz R, Bryngelsson T (2017) Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) germplasm from Southern Africa as revealed by microsatellite markers and agro-morphological traits. Genet Resour Crop Evol 64(3):599–610. https://doi.org/10.1007/s10722-016-0388-x
Muchira N, Ngugi K, Wamalwa LN, Avosa M, Chepkorir W, Manyasa E, Nyamongo D, Odeny DA (2021) Genotypic variation in cultivated and wild sorghum genotypes in response to Striga hermonthica infestation. Front Plant Sci. https://doi.org/10.3389/fpls.2021.671984
Muraya MM, de Villiers S, Parzies HK, Mutegi E, Sagnard F, Kanyenji BM, Kiambi D, Geiger HH (2011) Genetic structure and diversity of wild sorghum populations (Sorghum spp.) from different eco-geographical regions of Kenya. Theor Appl Genet 123(4):571–583. https://doi.org/10.1007/S00122-011-1608-6
Muui CW, Muasya R, Kirubi D (2013) Baseline survey on factors affecting sorghum production and use in eastern Kenya. Afr J Food Agric Nutr Dev 13(1):7339–7353. https://doi.org/10.4314/ajfand.v13i1
Muui C, Nguluu S, Kambura AK, Peter K (2019) Sorghum landraces production practices in Nyanza, Coast and Eastern regions, Kenya. J Econ Sustain Dev. https://doi.org/10.7176/jesd/10-10-16
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32(2):261–285. https://doi.org/10.1080/13102818.2017.1400401
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292. https://doi.org/10.1086/282771
Ngugi K, Onyango CM (2012) Analysis of the molecular diversity of Kenyan sorghum germplasm using microsatellites. J Crop Sci Biotechnol 15(3):189–194. https://doi.org/10.1007/S12892-012-0009-y
Ng’uni D, Geleta M, Bryngelsson T (2011) Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) accessions of Zambia as revealed by simple sequence repeats (SSR). Hereditas 148(2):52–62. https://doi.org/10.1111/J.1601-5223.2011.02208.x
Ogeto RM, Cheruiyot E, Mshenga P, Onyari CN (2013) Sorghum production for food security: a socio-economic analysis of sorghum production in Nakuru County, Kenya. Afr J Agric Res 8(47):6055–6067. https://doi.org/10.5897/ajar12.2123
Paradis E, Schliep K (2019) APE 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35(3):526–528. https://doi.org/10.1093/bioinformatics/bty633
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. https://doi.org/10.1093/bioinformatics/btg412
Poehlman JM (1987) Breeding sorghum. In: Breeding Field Crops. pp. 508–588.
Raatz B, Mukankusi C, Lobaton JD, Male A, Chisale V, Amsalu B, Fourie D, Mukamuhirwa F, Muimui K, Mutari B, Nchimbi-Msolla S, Nkalubo S, Tumsa K, Chirwa R, Maredia MK, He C (2019) Analyses of African common bean (Phaseolus vulgaris L.) germplasm using a SNP fingerprinting platform: diversity, quality control and molecular breeding. Genet Resour Crop Evol 66(3):707–722. https://doi.org/10.1007/s10722-019-00746-0
Salem KFM, Sallam A (2016) Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. C R Biol 339(1):1–9. https://doi.org/10.1016/j.crvi.2015.11.003
Smith CW, Frederiksen RA (2000) Sorghum: Origin, history, technology and production. Vol. 2. John Wiley & Sons, New York
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. https://doi.org/10.1111/J.1558-5646.1984.tb05657.x
Wondimu Z, Dong H, Paterson AH, Worku W, Bantte K (2021) Genetic diversity, population structure, and selection signature in Ethiopian sorghum [Sorghum bicolor L. (Moench)] germplasm. G3. https://doi.org/10.1093/g3journal/jkab087
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19(3):395–420. https://doi.org/10.1111/j.1558-5646.1965.tb01731.x
Wright S (1978) Variability within and among natural populations. In: Evolution and the genetics of populations. University of Chicago Press, Chicago
Funding
This research received funding from the National Research Fund (NRF) Kenya, under Grant Agreement No. NRF/1/MMC/244. The authors are thankful to the International Livestock Research Institute (ILRI), South Eastern Kenya University, Kenyatta University, and the University of Nairobi for facilitating the research, technical support, and exchange of information.
Author information
Authors and Affiliations
Contributions
The authors contributed to the study as follows: conceptualization: CWM, RMM, SN, and WK; study design: WK; data collection: PM; analysis and investigation: PM and WK; writing of draft and final manuscript: PM; final manuscript: all the authors reviewed and approved the final manuscript; funding acquisition: CWM, RMM, and SN; and supervision: LNW, FN, and CWM.
Corresponding author
Ethics declarations
Competing Interests
The authors have no financial or non-financial competing conflict of interest relevant to this article.
Additional information
Handling editor: Jody Hey.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mudaki, P., Wamalwa, L.N., Muui, C.W. et al. Genetic Diversity and Population Structure of Sorghum (Sorghum bicolor (L.) Moench) Landraces Using DArTseq-Derived Single-Nucleotide Polymorphism (SNP) Markers. J Mol Evol 91, 552–561 (2023). https://doi.org/10.1007/s00239-023-10108-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-023-10108-1