Abstract
Anchote (Coccinia abyssinica (Lam.) Cogn.) is a perennial root crop belonging to Cucurbitaceae family. It is endemic to Ethiopia and distributed over wide range of agro-ecologies. For further improvement and efficient conservation of this crop, characterization of its genetic diversity and its pattern of distribution is a vitally important step. Expressed sequence tags-simple sequence repeats (EST-SSRs) markers were developed from publicly available watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] ESTs in the GenBank database. Among those novel markers, eight were polymorphic and subsequently used for genetic diversity and population structure analyses of 30 anchote accessions collected from western Ethiopia. A total of 24 alleles were obtained across the eight polymorphic loci and 30 accessions that revealed moderate level of genetic diversity in this minor crop. Among the eight loci, locus CA_06 was the most informative with six alleles and polymorphic information content (PIC) of 0.76. The accessions showed about threefold variation in terms of genetic diversity, with expected heterozygosity (He) ranging from 0.15 (accession An) to 0.44 (accession Dg). Other accessions with higher genetic diversity include Ar and Gu (He = 0.43 and 0.41, respectively). Analysis of molecular variance (AMOVA) revealed that the variation within accessions and among accessions accounted for 84.7% and 15.3% of the total variation, respectively. The study revealed low but significant population differentiation in this crop with no clear pattern of population structure. The EST-SSR markers developed in this study are the first of their kind for anchote and can be used for characterization of its wider genetic resources for conservation and breeding purposes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anchote (Coccinia abyssinica (Lam.) Cogn.) is a perennial root crop that belongs to Cucurbitaceae family (Edwards et al. 1995; Hora et al. 1995). It is endemic to Ethiopia and distributed over a wide range of agro-ecologies adapting to altitudes as low as 550 m above sea level (masl) and as high as 2800 masl (Getahun 1973). The plant grows well in areas where annual rainfall is between 950 and 2000 mm (Getahun 1973; Westphal 1974). This species can reproduce both sexually and asexually through their root tubers. Although anchote is monoecious, the main mode of sexual reproduction is through outcrossing due to protandry (Edwards et al. 1995). Asexually, it undergoes an annual cycle in which its herbaceous shoots die out and then new shoots emerge at the onset of rainy season from its “everlasting” rootstock. Sowing seeds is a preferred means of anchote cultivation as the crop produces hundreds of seeds per fruit (Hora et al. 1995; Wondimu et al. 2014).
Anchote tuber is rich in protein and calcium with low content of anti-nutritional factors, and hence highly recommended as human food (Hora et al. 1995). It has also been used as traditional medicine, in Ethiopia, to treat various illnesses such as Bone fracture, backache, displaced joints and other diseases such as gonorrhea, tuberculosis, and cancer (Dawit and Estifanos 1991; Gelmesa 2010). Even though anchote is economically, nutritionally, and medicinally valuable crop plant, there is limited information regarding its genetics, breeding, best agronomic practices and phylogeography to date. Recent studies on its nutritional contents (Desta 2011), morphological traits (Wondimu et al. 2014) and genetic diversity using ISSR markers (Bekele et al. 2014) revealed key information that can be used for its improvement. However, these studies targeted only limited geographical areas where this crop is currently grown in Ethiopia, and hence the sample size and geographic coverage in previous study did not fully represent the wider gene pool of anchote in the country.
In order to harness the nutritional values and other benefits of this crop, quantifying its genetic diversity is a primary step. At present, single nucleotide polymorphism (SNP) markers are the marker of choice as they have genome-wide coverage and abundance in genic regions of most studied crops. However, such molecular markers require significant investment in terms of resources and time and hence are not an immediate option for orphan crop like anchote. Alternatively, expressed sequence tags-simple sequence repeats (EST-SSRs) are the second best option, as they are quick and cheaper to develop and use (Ellis et al. 2006). EST-SSRs markers have attributes, such as ease to develop, multi-allelic, transferable across genera and are from expressed section of the genome. They are also among markers of choice for QTL analysis to identify genes driving traits of agronomic interest (Ellis and Burke 2007). Therefore, developing such markers for anchote, would lead to enhanced understanding of its genome in general. In particular, such molecular tool can guide in-situ and ex-situ conservation efforts in Ethiopia and also play a role in breeding new varieties. Therefore, the present study was aimed at developing new EST-SSR markers for anchote and uses them for assessing the genetic diversity and population structure of anchote accessions grown in Western Ethiopia where this crop is staple food.
Materials and methods
Plant material
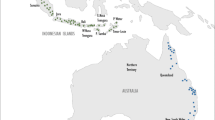
Seed samples were collected from anchote accessions in 2013 from areas where the plant exists in nature and where it is widely cultivated by small-scale farmers (Fig. 1). Thirty accessions, each represented by five individual plants, were selected based on their geographic distribution in the sampling area and their seeds were planted at Holeta Agricultural Research Center, Ethiopian Institute of Agricultural Research, located 41.7 km west of Addis Ababa during the year 2013/14 crop growing season. Three weeks after planting, young and healthy leaves were separately sampled from each seedling representing the 30 accessions. Leaf samples were kept in a plastic zipper containing sufficient silica gel for efficient desiccation and stored at room temperature until DNA extraction was conducted.
DNA extraction
Each silica gel dried leaf sample was transferred to a 2 ml eppendorf tube containing two glass beads and was ground to fine powder using Mixer Mill MM 400 (Retsch GmbH, Germany). The DNA extraction from the ground samples was done following protocol described in Geleta et al. (2012). The quality and quantity of extracted DNA was assessed by conducting agarose gel electrophoresis (1.5% (w/v) and NanoDrop measurement. The genomic DNA extracted from four of the samples was of poor quality and consequently discarded. Hence, genomic DNA from 146 individual plants representing the 30 accessions was used in this study (Table 1).
EST-SSRs screening and PCR
Watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] expressed sequence tags were downloaded from National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/nuccore). Then, WebSat (Martins et al. 2009), a web software for microsatellite marker development, was used to identify ESTs containing simple sequence repeats (di, tri, tetra, penta and hexa repeats). This was followed by using Primer3 (Rozen et al. 2000), an online primer designing program, for designing primer-pairs targeting the SSR containing regions of the ESTs. At the end, forty-seven primer-pairs were successfully designed and tested to determine their potential in amplifying the homologous DNA regions in anchote genome. Five, randomly selected, genomic DNA of anchote accessions were used to test these primer-pairs.
The total reaction volume for the amplification of the EST-SSRs was 25 μl composed of 2.5 μl of 10 × PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl, 1.25 mM MgCl2), 1.5 μl of 25 mM MgCl2, 0.3 μl of 25 mM dNTPs, 0.75 μl of 10 μM of each of forward and reverse primers, 1.0 U (0.2 μl) of Dream Taq DNA polymerase (Sigma, Germany), 2.5 μl of 10 ng template DNA, and 16.5 μl Millipore water. PCR amplification was performed in 96-well plates using Thermal Cycler (S 1000TM) machine with the following touchdown PCR conditions: Initial 95 °C for 3 min preheating, nine cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 45 s and primer extension at 72 °C for 45 s. The later three steps were repeated nine times by reducing the annealing temperature by 1 °C for each subsequent touchdown cycle. This was followed by 30 cycles of 94 °C denaturing for 30 s, 48 °C primer annealing for 45 s and 72 °C primer extension for 45 s. At the end of these cycles, additional 3 min primer extension was done at 72 °C. To check quality and size of amplifications, sub-sample, from each PCR reactions were loaded to ethidium bromide containing 1.5% agarose gel and electrophoresed using 1xTAE buffer. The gels were photographed using gel documentation system and the fragment sizes were compared with 50 bp DNA ladder (GeneRuler™, Fermentas Life Sciences), which was used as molecular size standard.
Out of the 47 EST-SSRs primer-pairs tested, 12 primer-pairs amplified a maximum of two DNA fragments of expected size per sample and hence were selected for use in this study. The description of these 12 primer-pairs is provided in Table 2. The forward primers were 5′-end labeled with carboxyfluorescein (6-FAM™) or hexachlorofluorescein (HEX™) fluorescent dyes, whereas the reverse primers were tailed with GCTTCT to reduce polyadenylation and improve genotyping as described in Ballard et al. (2002). These 12 labeled primer-pairs were used for PCR amplifications of the DNA samples extracted from the 146 individuals representing the 30 accessions (Table 2).
The amplified products were multiplexed into panels based on the fragment size differences and the type of fluorescent dye of the forward primers. This was followed by capillary gel electrophoresis using ABI Prism® 3730xl genetic analyzer (Applied Biosystems). The peak identification and fragment size determination, based on the Genescan-500 LIZ internal size standard, were done using GeneMarker ver. 2.4.0 (SoftGenetics).
Data analyses
Various genetic diversity parameters were analyzed using different software. Observed number of allele (Na), effective number of alleles (Ne), percentage of polymorphic loci (%PL), Shannon–Weaver diversity index (I), observed heterozygosity (Ho), expected heterozygosity (He) and gene flow (Nm) were analyzed using POPGENE ver. 32 software (Yeh and Yang 999). DARwin ver. 6.0.112 software (2014) was used for cluster analysis of the 30 accessions using neighbor joining method. POWERMARKER ver. 3.25 software (Liu and Muse 2005) was used to calculate polymorphic information contents (PIC) for each locus (Anderson et al. 1993). Analysis of molecular variance (AMOVA) was calculated using ARLEQUIN ver. 3.01 software (Excoffier et al. 2005). HP-Rare ver. 1.0 software was used to calculate allelic richness and private allele Kalinowski (2005), allele frequency per locus as well as gene diversity per locus and per population. Excel add-ins (GENAIEX) ver.6.502 software Peakall and Smouse (2004) was used for pair-wise population differentiation, and pair-wise genetic distance.
The population structure analysis by clustering method and determination of optimum number of clusters (K) were conducted using STRUCTURE ver. 2.3.4 software (Pritchard et al. 2000) using admixture model (Gilbert et al. 2012). The STRUCTURE analysis was conducted for K = 2 to K = 25 with burn-in period of 200,000 and number of Markov Chain Mont Carol (MCMC) replications of 200,000, and 10 runs at each K. Optimum K value (maximum ΔK), estimation of different genetic groups, was determined by the STRUCTURE HARVESTER software according to Evanno et al. (2005). Cluster alignment across the replicates was done using CLUMPP software (Jakobsson and Rosenberg 2007) and the population clusters were visualized by DISTRUCT software (Rosenberg 2004).
Results
In present study twelve EST-SSR markers were developed for anchote of which nine were di-nucleotide repeats and the rest three were tri-nucleotide repeats (Table 2). Four of these loci (CA_03, CA_04, CA_05 and CA_12) were monomorphic (Table 2). The other eight loci showed different levels of polymorphism and their data was used for genetic diversity and population structure analyses of the 30 accessions. A total of 24 alleles were identified across the 146 samples and the eight loci (Table 3). The observed number of alleles per locus (Na) ranged from two to six with an average of three alleles per locus. The effective number of allele (Ne) ranged from 1.07 (locus CA_08) to 4.70 (locus AC_06) with an average of 1.90 (Table 4). The frequency of the alleles across all accessions varied from 0.01 (Alleles A and D of locus CA_01) to 0.97 (allele B of locus CA_08) (Table 3). Locus CA_06 is the most polymorphic loci with six alleles and polymorphic information content (PIC) of 0.76, whereas the lowest PIC (0.06) was recorded in locus CA_08. The highest (1.64) and lowest (0.14) Shannon’s information index (I) were recorded for loci CA_06 and CA_08, respectively. The eight loci have a mean PIC and I values of 0.31 and 0.63, respectively (Table 4). Among the loci with only two alleles, locus CA_11 is the most informative with PIC value of 0.37. Among the eight loci, locus CA_06 and locus CA_08 are the highest and lowest values (Table 4).
Gene diversity (He) estimated across the accessions for each locus varied from 0.06 (locus CA_08) to 0.79 (locus CA_06) with a mean of 0.37, whereas observed heterozygosity (Ho) varied from 0.01 (locus CA_08) to 0.84 (locus CA_11) with a mean of 0.35 (Table 5). Locus CA_01, CA_07, and CA_08 indicated significant inbreeding of individuals in each accession (FIS) whereas all loci, except CA_10 and CA_11, showed significant inbreeding of individual genotypes when compared to random association of alleles in the whole population (FIT). CA_02, CA_06 and CA_08 are major contributing loci for significant population differentiations (FST). The mean total (Ht), within accessions (Hs) and among accessions (DST) gene diversity were 0.37, 0.35 and 0.02, respectively. The overall average population differentiation (FST) and gene flow (Nm) were 0.11 and 2.06 (Table 5).
Gene diversity across the loci for each accession varied from 0.15 (accession An) to 0.44 (accession Dg). Other accessions with a relatively high diversity include Ar (0.43), Gm (0.41), Gu (0.41), Ac (0.40) and Sy (0.40) (Table 6). The average allelic richness of the accessions varied from 1.38 (accession An) to 2.38 (accession Ar) (Table 6). Accessions with second highest allelic richness (AR = 2.13) are Dw, Gm, Nj, Dg, Ld, Ac, and Bt.
A minimum frequency of major allele (FMA) observed was 0.2 for Ld accession by locus CA-10. The maximum allelic richness (AR) scored was 5.0 for both Ld and Bt accessions by the same locus, CA-06. Average gene diversity (He) for the 30 anchote accessions for each of the eight polymorphic loci as well as their mean values are presented in Supplementary Table 1.
The pair-wise Nei’s standard genetic distance between the accessions varied from 0.02 (accessions Ss vs Sh) to 0.48 (accessions Gu vs Ds) with the overall mean genetic distance of 0.13 (Supplementary Table 2). 12.9% of the accession-pairs had genetic distance of above 0.2 between them whereas 34.9% pairs had genetic distance of below 0.1 between them (data not shown). Accession Ds is the most differentiated with a mean genetic distance of 0.30 from the other accessions whereas accessions Ar and Ac are the least differentiated with a mean genetic distance of 0.09. The analysis of pair-wise FST revealed that 12.9% accession-pairs were significantly differentiated (Supplementary Table 2).
Analysis of molecular variance (AMOVA) of the genotypic data of the 30 accessions partitioned the total variance into among individuals within accessions and among accessions components which accounted for 84.65% and 15.35% of the total variance, respectively. The analysis revealed a significant population differentiation with FST value of 0.15 (P < 0.001) (Table 7). Furthermore, the genetic relationship between the 30 accessions was determined through Neighbor-joining cluster analysis based on Nei’s standard genetic distance between the accessions. The cluster analysis revealed three major clusters, with cluster-I, II and III comprising 25, 2 and 3 accessions, respectively (Fig. 2), but fails to show any clear association with geographic origin. The optimal number of genetic clusters (K) revealed through admixture model-based population structure analysis was four (Fig. 3a). Hence, the 146 individuals of the 30 accessions most likely originated from four genetic populations. The analysis clearly suggested low differentiation between the accessions, as all accessions have alleles originated from the four genetic populations (clusters) as shown by graphical representation of the population genetic structure of the 30 accessions at K = 4 (Fig. 3b).
a ΔK plot showing its maximum value at K = 4 suggesting the optimal number of genetic clusters (populations) of four; and b CLUMPAK plot (K = 4) of the 30 anchote accessions based on the STRUCTURE and STRUCTURE Harvester out puts. Each color represents a different cluster and black vertical lines separate the accessions, and accession codes are shown under the plot
Discussion
Anchote has a good potential to share the burden of cereals and other crops as additional calorie source in Ethiopia. The fact that anchote is a drought tolerant tuber crop (Getahun 1973), makes it ideal candidate for the current erratic climatic conditions characterized by frequent droughts. However, despite its attributes in terms of economy, nutrition and resilience, it has yet to receive research attention and only semi-domesticated (Dawit and Estifanos 1991; Hora 1995; Gelmesa 2010). In the present study, a total of 12 EST-SSR markers were developed from publicly available EST sequences of watermelon. The fact that these markers were able to amplify expected size fragments in anchote, opens the door of adopting other genomic tools such as SNPs/indels developed in related species, like watermelon, for anchote gene pool characterization and improvement. In addition, these newly developed EST-SSR markers can be a vital tool for taxonomic status determination of the species and subspecies of the genus Coccinia.
Among the newly developed EST-SSR markers, four were monomorphic across 146 individuals included in study, hence excluded from further diversity analyses. These monomorphic loci may turn out to be polymorphic if additional anchote accessions from wider geographic areas were to be included, because analysis of a wider gene pool of crop species facilitates the identification of rare alleles (Kalinowski 2004). Rare alleles within genes of significant functions are usually related to out-breeding species, like anchote, where they occur in heterozygous form; otherwise can be deleterious in their homozygote state (Frankham 2002). Hence, further study is required to elucidate the significance of these loci in population genetics analyses of anchote.
The two commonly occurring types of microsatellites, in flowering plant species, are di-nucleotide and tri-nucleotide (Simko 2009; Cavagnaro et al. 2010; Wen et al. 2010; Dillon, et al. 2014). In the present study, di, tri, tetra, penta and hexanucleotide repeat EST-SSRs were included in the initially targeted 47 loci although most of them were di and trinucleotide repeat SSRs. However, only the dinucleotide and trinucleotide repeat SSRs successful in amplifying homologous regions in anchote (Table 1), suggesting the higher transferability of these two groups of SSRs than the other groups. However, changes in repeat number of di-nucleotide repeat SSRs causes frame shift mutation that may alter the associated protein and/or function (Metzgar et al. 2000). Interestingly, the highest allelic richness in the present study was observed for locus CA_06 which is a di-nucleotide repeat. The other polymorphic loci were also moderately informative, with locus CA_01 (trinucleotide repeat) being the second most polymorphic locus with four alleles. In general, the maximum number of alleles per locus (EST-SSR marker) was six, which is comparable with what was observed in related species like Cucumis sativus (Kong et al. 2006), Cucurbitaceae species (Reddy 2009) and Cucurbita pepo (Mao et al. 2014).
The moderate genetic diversity observed in this study indicates that strong selection pressure; probably farmers’ intervention to improve yield and related traits may have caused reduced polymorphism. A similarly narrow genetic base in cultivated cultivars of watermelon was observed when genotyped with similar or other types of molecular markers (Wang 2011). Therefore, in the future breeding efforts, accessions with highest genetic diversity among the 30 accessions such as Dg, Ar and Gm should be considered for diversifying the genetic base of cultivated varieties and also to incorporate resilient genes capable of withstanding different abiotic stresses. On the contrary, An, Dh, Gc were the accessions with the lowest genetic diversity observed, which might be inferred to recent adaptation of anchote to the areas where these accessions were collected (founder effect) or recent bottleneck (Hawks et al. 2000).
The highest values of fixation indices for inbreeding (FIS and FIT) were recorded for loci CA_08 and CA_07 (Table 4). Theses loci have higher heterozygote deficiency, because for most populations the alleles of these loci were less than two. Such loci should be given further research attention as they might be linked to or located at vital genes that minimize recombination frequency in order to prevent loss of vital function. Contrary to these loci, locus CA_11 was represented by excess heterozygosity which suggest heterozygote advantage at that locus. Multi-allelic loci are of greater value for poorly studied crops like anchote as they can serve as molecular barcodes for variety/accession identification or can play a role in efficient conservation and management of anchote germplasms in Ethiopia and beyond. Hence, loci CA_06 and CA_01 should get priority for these purposes, as they have higher PIC than other loci. In this study, low but significant differentiation was obtained between the accessions (FST = 0.15; P < 0.001) with 54.2% of the accession-pairs showing significant differentiation. Similar study using inter simple sequence (ISSR) markers revealed higher population differentiation (FST = 0.49) (Bekele et al. 2014). Lower population differentiation and reduced genetic diversity are expected when EST-SSRs are used instead of neutral markers such as ISSR, as mutation rate in EST-SSRs is relatively low (Serra et al. 2007). Hence, significantly differentiated accessions need to be given priority both for conservation and breeding purposes. The low population differentiation is attributable to the moderate level of gene flow between anchote populations similar with other studies (Wickert et al. 2012). In future germplasm collecting missions, districts from which more diverse populations were obtained should be given priority in order to capture further diversity. Analysis of molecular variance revealed that about 85% of the total variation accounted for the variation within accessions. This is probably due to direct germplasm exchange between farmers of different districts, and through local market system as well as pollen flow between anchote populations.
The genetic distance between populations did not follow the geographic distance between the sampling sites of the accessions. The neighbor-joining cluster analysis also clearly showed complex patter of clustering that is not in line with the geographic origin of the accessions. The population structure analysis revealed high level of admixture between the accessions. The analysis also revealed not much differences between the genotypes in terms of the likelihood of their membership in each of the genetic populations, indicating lack of clear population structure in anchote. The anchote accessions included in the present study were collected from areas where the local communities share similar socio-economic and food culture, which might have resulted lower than expected differentiation/diversity between the accessions and lack of clear population structure. It would be interesting to analyze anchote germplasm representing local communities with different socioeconomic and food culture in the future studies.
Conclusions
The development of novel EST-SSR markers, and their application for the analyses of genetic diversity, is the first of its kind in anchote. Although the number of markers developed in this study is few, they have an important role to play in characterization of anchote genetic resources for conservation and breeding purposes. The result of this study also serves as additional evidence in supporting the across-species transferability, within a family, of a good proportion of EST-SSR markers. The present study revealed moderate level of genetic diversity in anchote accessions and suggested potential hotspots for conservation. However, further research including more germplasms and molecular markers is required in order to have a clearer pattern about its population genetics as well as for its improvement through breeding. Hence, this study serves as a motivation for researchers to develop genomic tools for this vital but orphan crop so that modern breeding tools such marker assisted selection (MAS) and genomic selection (GS) can be applied, for accelerated delivery of high yielding and “climate smart” anochte varieties with wider adoption which in turn contributes towards food and nutrition security in Ethiopia and Eastern Africa.
Availability of data and materials
The authors declare that the data supporting the findings of this study are available within the article.
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- AR:
-
Allelic richness
- EST:
-
Expressed sequence tags
- FMA:
-
Frequency of major allele
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
References
Anderson J, Churchill G, Autrique J, Tanksley S, Sorrells M (1993) Optimizing parental selection for genetic linkage maps. Genetics 36:181–186
Ballard LW, Adams PS, Bao Y, Bartely D, Bintzler D, Kasch L, Petukhova L, Rosato C (2002) Strategies for genotyping: electiveness of tailing primers to increase accuracy in short tandem repeat determinations. J Biomol Technol 13:20–29
Bekele A, Feyissa T, Tesfaye K (2014) Genetic diversity of anchote (Coccinia abyssinica (Lam.) Cogn.) from Ethiopia as revealed by ISSR markers. Gen Res Crop Evol 61:707–719
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumissativus L.). BMC Genom 11:1–18
Dawit A, Estifanos H (1991) Plants as a primary source of drugs in the traditional health practices of Ethiopia. In: Engles JMM, Hawkes JG, Worede M (eds) Plant genetic resources of Ethiopia. Cambridge University Press, Cambridge
Desta F (2011) Phenotypic and nutritional characterization of anchote [Coccinia abyssinica (Lam.) Cogn] accessions of Ethiopia. M.Sc. thesis, Jimma University
Dillon NL, Innes DJ, Bally ISE, Wright CL, Devitt LC, Dietzgen RG (2014) Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Mangifera indica L.). Diversity 6:72–87
Edwards S, Tadesse M, Hedberg I (1995) Flora of Ethiopia and Eritrea. National Herbarium, Addis Ababa University and Uppsala University, Addis Ababa
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Ellis JS, Knight ME, Darvill B, Goulson D (2006) Extremely low effective population sizes, genetic structuring, and reduced genetic diversity in a threatened umblebee species, Bombus sylvarum (Hymenoptera: Apidae). Mol Ecol 15:4375–4386
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Biol 1:47–50
Frankham R, Ballou JD, Briscoe DA, Mclnness KH (2002) Introduction to conservation genetics. Cambridge University Press, New York
Geleta M, Herrera I, Monz A, Bryngelsson T (2012) Genetic Diversity of arabica coffee (Coffea arabica L.) in Nicaragua as estimated by simple sequence repeat markers. Sci World J 2012:1–11
Gelmesa D (2010) Shifting to alternative food source: potential to overcome Ethiopians’ malnutrition and poverty problems. In: Innovation and sustainable development in agriculture and food (ISDA), Montpellier-France, pp 1–10
Getahun A (1973) Developmental anatomy of tubers of anchote: a potential dry land crop. Acta Hortic Tech Commun ISHS 33:51–64
Gilbert KJ, Andrew RL, Bock DG, Franklin MT, Kane NC, Moore JS, Moyers BT, Renaut S, Rennison DJ, Veen T, Vines TH (2012) Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program STRUCTURE. Mol Ecol 21(20):4925–4930
Hawks J, Hunley K, Lee SH, Wolpoff M (2000) Population bottlenecks and pleistocene human evolution. Mol Biol Evol 17(1):2–22
Hora A, Edwards S, Giday M, Tesfaye Y (1995) Anchote an endemic tuber crop. Artistic Printing Press Enterprise, Addis Ababa
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(18):1–6
Kalinowski ST (2004) Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv Genet 5:39–543
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kong Q, Xiang C, Yu Z (2006) Development of EST-SSRs in Cucumis sativus from sequence database. Mol Ecol Notes 6:1234–1236
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mao W, Xu S, Ye S, Wang H, Mo GX, Gong X, Y, (2014) Development of 19 transferable Cucurbita pepo EST-SSR markers for the study of population structure and genetic diversity in pumpkin (Cucurbita moschata). POJ 7(5):345–352
Martins WS, Lucas DCS, NevesBertioli KFDJ (2009) WebSat-A web software for microsatellite marker development. Bioinformation 3(6):282–283
Metzgar D, Bytof J, Wills C (2000) Selection against frame shift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Peakall PR, Smouse PP (2004) GenAlEx 6.5: genetic analysis in excel. population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Redd BU (2009) Cladistic analyses of a few members of Cucurbitaceae using rbcL nucleotide and amino acid sequences. Int J Biol Res 1:58–64
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Meth Mol Biol 132:365–386
Serra IA, Procaaaini G, Intrieri MC, Migliaccio M, Mazzuca S, Innocenti AM (2007) Comparison of ISSR and SSR markers for analysis of genetic diversity in the seagrass Posidonia oceanica. Mar Ecol Prog Ser 338:71–79
Simko I (2009) Development of EST-SSR markers for the study of population structure in lettuce (Lactuca sativa L.). J Her 100:256–262
Wang JH, Kang J, Son B, Kim K, Park Y (2011) Genetic diversity in watermelon cultivars and related species based on AFLPs and EST-SSRs. Not Bot Hortic Agrobiol 39(2):285–292
Wen W, Wang H, Xia Z, Zou M, Lu C, Wang W (2010) Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha curcas L. BMC Res Notes 3(42):1–8
Westphal E (1974) Pulses in Ethiopia, their taxonomy and agricultural significance. Doctoral thesis, Wageningen: Centre for Agricultural Publishing and Documentation
Wickert E, de Goes A, de Souza A, Lemos EG (2012) Genetic diversity and population differentiation of the causal agent of citrus black spot in Brazil. Sci World J 2012:1–14. https://doi.org/10.1100/2012/368286
Wondimu T, Alamerew S, Ayana A, Garedew W (2014) Genetic diversity analysis among anchote (Coccinia abyssinica) accessions in western Ethiopia. Int J Agric Res 9:149–157
Yeh FC, Yang R (1999) Popgene Version 1.31. Microsoft Window based freeware for population genetic analysis. University of Alberta and Tim Boyle, Centre for International Forestry Research, Alberta, Canada, vol 1, pp 1–28
Acknowledgement
Wollega University partially financed this work. We thank Addis Ababa University (Microbial, Cellular and Molecular Biology Department) and Swedish University of Agricultural Sciences (Department of Plant Breeding) for their collaboration to facilitate this research; Holeta Agricultural Research Center is acknowledged for allowing us to grow anchote accessions used for this study at their experimental site.
Funding
This research work is financed by the Swedish Research Council (VR) through Swedish Research Link project grant and Addis Ababa University’s Thematic Research Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Not applicable.
Consent to publish
We all agree to publish this work.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Serbessa Tolera, B., Dagne Woldegebriel, K., Teshome Gari, A. et al. Analyses of genetic diversity and population structure of anchote (Coccinia abyssinica (Lam.) Cogn.) using newly developed EST-SSR markers. Genet Resour Crop Evol 68, 2337–2350 (2021). https://doi.org/10.1007/s10722-021-01132-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01132-5