Abstract
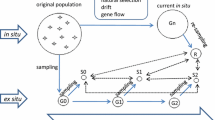
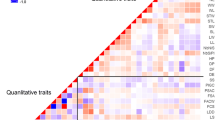
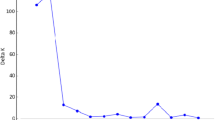
Maintaining germplasm genetic integrity is a key objective of long-term ex situ conservation. Periodic regeneration, performed on limited plots with small number of individuals, increases the risk of genetic drift and genetic diversity changes. In this study, six accessions of white flowered, dry seed pea varieties (Pisum sativum L. subsp. sativum var. sativum): Bohatýr, Klatovský zelený, Hanák, Moravský hrotovický krajový, Raman and Viktoria-75 and four accessions of colour flowered, fodder pea (P. sativum subsp. sativum var. arvense (L.) Poiret: Arvika, Český banán, Moravská krajová and Niké, representing Czechoslovak varieties and landraces, bred over the last 40–80 years, were analyzed using ten microsatellite locus specific markers. Each accession was represented by 20 individual seeds of two temporally different samples, spanning the period of 20 or 40 years. Together with intra-accession variation (except of cv. Hanák), evidence of genetic changes, e.g. differences in allele frequencies as well as genetic composition of sample, was detected in six out of ten accessions (Arvika, Bohatýr, Český banán, Moravský hrotovický krajový, Moravská krajová and Raman). Evidence of genetic erosion was found in three accessions (Český banán, Moravský hrotovický krajový and Raman), while in another three (Arvika, Bohatýr and Moravská krajová) the level of diversity was found to have increased. Moreover in three samples of Bohatýr (2004) and Klatovský zelený (1963 and 2004), low levels of heterozygosity was detected. These results demonstrate that in pea, a self-pollinating and highly homozygous plant, the danger of the loss of genetic integrity exists. These findings are significant for long-term ex situ germplasm management.

Similar content being viewed by others
References
Ambrose MJ (1995) From near east centre of origin the prized pea migrates throughout world. Diversity 11:118–119
Baranger A, Aubrey G, Aran G, Laine AL, Deniot G, Potier J, Burstin J (2004) Genetic diversity within Pisum sativum using protein and PCR-based markers. Theor Appl Genet 108:1309–1321
Baur E (1914) Die Bedeutung der primitiven Kulturrassen und der wilden Verwandten unserer Kulturpflanzen für die Pflanzenzüchtung. Jb Deut Landw Ges 29:104–109
Börner A, Chebotar C, Korzun V (2000) Molecular characterization of the genetic integrity of wheat (Triticum aestivum L.) germplasm after long-term maintenance. Theor Appl Genet 100:494–497
Breese EL (1989) Regeneration and multiplication of germplasm resources in seed genebanks: the scientific background. International Board for Plant Genetic Resources, Rome
Brown AHD (1989) Core collections: a practical approach to genetic resources management. Genome 31:818–824
Brown AHD, Brubaker CL, Grace JP (1997) Regeneration of germplasm samples: wild versus cultivated plant species. Crop Sci 37:7–13
Chebotar S, Roder MS, Korzun V, Borner A (2002) Genetic integrity of ex situ genebank collections. Cell Mol Biol Lett 7:437–444
Chwedorzewska KJ, Bednarek PT, Puchalski J, Krajewski P (2002) AFLP-profiling of long term stored and regenerated rye genebank samples. Cell Mol Biol Lett 7:457–463
Dostálová R, Seidenglanz M, Griga M (2005) Simulation and assessment of possible environmental risks associated with release of genetically modified peas (Pisum sativum L.) into environment in Central Europe. Czech J Genet Plant Breed 41:51–63
Enjabert J, Goldringer I, Paillard S, Brabant P (1999) Molecular markers to study genetic drift and selection in wheat populations. J Exp Botany 50:283–290
Esquinas-Alcázar J (2005) Protecting crop genetic diversity for food security: political, ethical and technical challenges. Nat Rev Genet 6:946–953
FAOSTAT (2007) Databases Query, http://faostat.fao.org/
Figliuolo G, Mazzeo M, Greco I (2007) Temporal variation of diversity in Italian durum wheat germplasm. Genet Resour Crop Evol 54:615–626
Frankel OH, Brown AHD (1984) Current plant genetic resources—a critical appraisal. In: Chopra VL, Joshi BC, Sharma RP, Bansal HC (eds) Genetics: new Frontiers, vol IV. Oxford & IBH Publ, New Delhi, pp 1–11
Genebank Standards (1994) Food and Agriculture Organization of the United Nations, Rome, International Plant Genetic Resources Institute, Rome
Goudet J (1995) FSTAT (ver.1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Griga M, Smýkal P, Dostálová R, Seidenglanz M (2008) Field assessment of outcrossing rate in pea (Pisum sativum L.), a cleistogamous crop, as a background for release of genetically modified pea into environment. In: 10th international symposium on biosafety of genetically modified organisms, symposium handbook: biosafety research of GMOs: past achievements and future challenges. Wellington, New Zealand, p 110
Gugerli F, Brodbeck S, Holderegger R (2008) Insertions–deletions in a microsatellite flanking region may be resolved by variation in stuttering patterns. Plant Mol Biol Rep 26:255–262
Horáček J, Griga M, Smýkal P, Hýbl M (2009) Effect of environmental and genetic factors on the stability of pea (Pisum sativum L.) isozyme and DNA markers. Czech J Genet Plant Breed 45:57–71
ICRISAT (1995) Consultation meeting on the regeneration of seed crops and their wild relatives. ICRISAT, Hyderabad, India
Jing RC, Knox MR, Lee JM, Vershinin AV, Ambrose M, Ellis THN, Flavell AJ (2005) Insertional polymorphism and antiquity of PDR1 retrotransposon insertions in Pisum species. Genetics 171:741–752
Le Clerc V, Briard M, Granger J, Delettre J (2003) Genebank biodiversity assessments regarding optimal sample size and seed harvesting techniques for the regeneration of carrot accessions. Biodiv Conserv 12:2227–2336
Lehmann CO, Mansfeld R (1957) Zur Technik der Sortimentserhaltung. Die Kulturpflanze 5:108–138
Lia VV, Bracco M, Gottlieb AM, Poggio L, Confalonieri VA (2007) Complex mutational patterns and size homoplasy at maize microsatellite loci. Theor Appl Genet 115:981–991
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G et al (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Malysheva-Otto L, Ganal MW, Law JR, Reeves JC, Röder MS (2007) Temporal trends of genetic diversity in European barley cultivars (Hordeum vulgare L.). Mol Breed 20:309–322
Mendel JG (1866) Versuche über Pflanzenhybriden. Verhandlungen des naturforschenden Vereines in Brünn, Bd. IV für das Jahr 1865
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Otto SP, Whitlock MC (1997) The probability of fixation in populations of changing size. Genetics 146:723–733
Parzies HK, Spoor W, Ennos RA (2000) Genetic diversity of barley landrace accessions (Hordeum vulgare ssp. vulgare) conserved for different length of time in ex situ gene banks. Heredity 84:476–486
Pavelková A, Moravec J, Hájek D, Bareš I, Sehnalová J (1986) Descriptor list of the genus Pisum L. RICP Prague—Ruzyně. Genové zdroje 32:46
Reynolds J, Weir BS, Cockerham C (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–776
Roberts EH, Ellis RH (1984) The implications of the deterioration of orthodox seeds during storage for genetic resources conservation. In: Holden JHW, Williams JT (eds) Crop genetic resources: conservation and evaluation. IBPGR, George Allen and Unwin Ltd. Rome, International Plant Genetic Resources Institute, Rome, pp 19–37
Russell JR, Ellis RP, Thomas WTB, Waugh R, Provan J, Booth A, Fuller J, Lawrence P, Young G, Powell W (2000) A retrospective analysis of spring barley germplasm development from ‘foundation genotypes’ to currently successful cultivars. Mol Breed 6:553–568
Sackville HNR, Chorlton KH (1997) Regeneration of accessions in seed collections: a decision guide. Handbook for genebanks No. 5. International Plant Genetic Resources Institute, Rome
Schindler F (1890) Welches Werthverhältniss besteht zwischen den Landrassen landwirthschaftlicher Kulturpflanzen und den sogenannten Züchtungsrassen ? Internationaler land- und forstwirtschaftlicher Congress zu Wien 1890. Section I: Landwirthschaft. Subsection: Pflanzenbau. Frage 5. Heft 13:19–24
Smýkal P (2006) Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J Appl Genet 47:221–230
Smýkal P, Horáček J, Dostálová R, Hýbl M (2008a) Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J Appl Genet 49:155–166
Smýkal P, Hýbl M, Corander J, Jarkovský J, Flavell A, Griga M (2008b) Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor Appl Genet 117:413–424
Smýkal P, Kalendar R, Ford R, Macas J, Griga M (2009) Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity 103:157–167
Soengas P, Cartea E, Lema M, Velasco P (2009) Effect of regeneration procedures on the genetic integrity of Brassica oleracea accessions. Mol Breed 23:389–395
Tanksley SD, McCouch SR (1997) Seed bank and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
UPOV-BMT: BMT/36/10. (2002) Progress report of the 36th session of the technical committee, the technical working parties and working group on biochemical and molecular techniques and DNA-profiling in particular. Geneva
van Hintum TJL, Van De Wiel CCM, Visser DL, Van Treuren R, Vosman B (2007) The distribution of genetic diversity in a Brassica oleracea gene bank collection related to the effects on diversity of regeneration, as measured with AFLPs. Theor Appl Genet 114:777–786
von Proskowetz E (1890) Welches Werthverhältniss besteht zwischen den Landrassen landwirthschaftlicher Kulturpflanzen und den sogenannten Züchtungsrassen ? Internationaler land- und forstwirtschaftlicher Congress zu Wien 1890. Section I: Landwirthschaft. Subsection: Pflanzenbau. Frage 5. Heft 13:3–18
Wright S (1965) The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution 19:395–420
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian J Botany 129:157
Zohary D, Hopf M (2000) Domestication of plants in the Old World. Oxford University Press, Oxford
Acknowledgments
This work was supported by the Ministry of Education of Czech Republic, MSM 2678424601 project. Excellent technical assistance of Mrs. E. Fialová, valuable discussion and manuscript improvement by Dr. C. Coyne, USDA-ARS Pullman and Dr. Mike Ambrose, JIC Norwich UK are greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jaroslava Cieslarová and Petr Smýkal authors contributed equally to the work.
Rights and permissions
About this article
Cite this article
Cieslarová, J., Smýkal, P., Dočkalová, Z. et al. Molecular evidence of genetic diversity changes in pea (Pisum sativum L.) germplasm after long-term maintenance. Genet Resour Crop Evol 58, 439–451 (2011). https://doi.org/10.1007/s10722-010-9591-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9591-3