Abstract
Aposematic organisms rely on their bright conspicuous coloration to communicate to potential predators that they are toxic and unpalatable. These aposematic phenotypes are strongly tied to survival and therefore make excellent opportunities to investigate the genetic underpinning of coloration. The genus Ranitomeya includes phenotypically diverse members of Neotropical aposematic poison frogs native to South America. Significant progress has been made in elucidating the molecular mechanisms responsible for aposematic coloration in poison frogs, which have paved the way for future studies to test hypotheses of the evolution of coloration across aposematic vertebrates. However, very little is known about whether these color related genes are under positive selection. We assembled transcriptomes from publicly available data reads sets for 9 different color morphs of poison frogs in the Ranitomeya genus that display bright conspicuous coloration (four morphs of R. imitator, two morphs of R. variabilis, two morphs of R. fantastica, one morph of R. summersi) to identify protein-coding genes responsible for color production that are under positive selection. Our results show that there are multiple genes under strong positive selection that are predicted to play roles in melanin synthesis (dct, tyrp1, irf4), iridophore development (fhl1), keratin metabolism (ovol1), pteridine synthesis (prps1, xdh), and carotenoid metabolism (adh1b, aldh2). The identification of positive selection affecting candidate color-pattern genes is consistent with the possibility that these genes mediate (in part) the molecular evolution of coloration. This may be attributed to aposematic phenotypes being directly tied to survival and reproduction in poison frogs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Visible color provides a unique window into fundamental aspects of ecology and evolution, and evolutionary biologists have long been focused on the morphological and molecular makeup of coloration in animals (Longley 1917; Fox 1936; Gray and McKinnon 2007). The diversity of color phenotypes is mainly attributed to selective pressures from both biotic (e.g., predator avoidance, sexual signaling) and abiotic (e.g., UV protection, thermoregulation) factors (Rudh and Qvarnström 2013), where color phenotypes displayed by animals range from cryptic to highly conspicuous. Bright coloration accompanied with contrasting patterns in nature is pervasive and plays vital roles in species recognition (Losos 1985), mate choice (Kokko et al. 2002), and predator avoidance (Ruxton et al. 2004). Coloration is strongly tied to survival and therefore plays a central role in various ecological and evolutionary processes.
In nature, coloration commonly evolves to promote prey survival. One common evolutionary solution is aposematism, in which species evolve bright conspicuous coloration to warn predators that they have a dangerous secondary defense and are therefore unpalatable (Harvey et al. 1982; Brown 2013; Cummings and Crothers 2013). Theoretical considerations suggest that closely related aposematic species under predator-imposed selection pressure would converge on a single conspicuous phenotype to accelerate predator learning (Müller 1879; Mallet and Joron 1999). For instance, phenomena such as Müllerian mimicry, in which two or more chemically defended species mimic each other’s visual warning signals (converging on a single phenotype) is ubiquitous in nature. However, geographic variation in the color and pattern of visual warning signals is common in aposematic species (Joron and Mallet 1998; Briolat et al. 2018, 2019).
Neotropical poison frogs in the family Dendrobatidae, native to Central and South America, provide an iconic example of aposematism. Roughly one-third of frogs in the family Dendrobatidae exhibit conspicuous coloration and are chemically defended by alkaloids sequestered from their diet (Harvey et al. 1982; Brown 2013; Cummings and Crothers 2013). Some species in this family possess high intraspecific diversity in color and pattern while others are monomorphic (Brown et al. 2011). Within the past decade, a considerable amount of effort has been put into identifying the genes responsible for color pattern production in aposematic poison frogs, particularly the mechanisms producing polytypism (Posso-Terranova and Andrés 2017; Stuckert et al. 2019, 2021, 2023; Rodríguez et al. 2020; Twomey et al. 2020a; Linderoth et al. 2023). However, very little is known concerning the patterns of molecular evolution of these genes.
The evolution of color phenotypes occurs under an array of selection pressures (Hebets and Papaj 2005; Caro et al. 2016; Cuthill et al. 2017). For instance, the diversity of vivid coloration found in Neotropical poison frogs is likely to be shaped by strong predator-induced selection pressures. However, identifying the natural predators of poison frogs is notably difficult and therefore remains understudied. Saporito et al. (2007) found that the majority of attacks on plasticine poison frog models are avian predators and that they actively avoided models with bright aposematic coloration. Avian predators have been considered to be the most important predators influencing the evolution of aposematic species (Chai 1986). It is worth mentioning that visual colors in aposematic species can also play vital roles in mate choice (Summers et al. 1999; Reynolds and Fitzpatrick 2007; Maan and Cummings 2009; Finkbeiner et al. 2014; Twomey et al. 2016; Yang et al. 2016). This evidence is consistent with the notion that bright conspicuous coloration accompanied with contrasting patterns is closely tied to reproductive success as well as survival, and therefore the genetic mechanisms responsible for such phenotypes are likely to be under positive selection.
In amphibians, the production of color and pattern is associated with three types of chromatophores (melanophores, iridophores, and xanthophores). Melanophores contain pigments called melanins, which are responsible for the production of black, brown, and dark green coloration (Duellman and Trueb 1994). Iridophores contain structural elements known as guanine platelets. Depending on their size, shape, and orientation, the platelets reflect different wavelengths of light and ultimately determine structural coloration (Ziegler 2003; Bagnara et al. 2007; Saenko et al. 2013). For example, long wavelengths occur when platelets are thick in size, resulting in an orange coloration, whereas short wavelengths occur when platelets are thin in size, resulting in a dark blue coloration (Twomey et al. 2020b). Xanthophores contain pigments composed of pteridine and carotenoids which are responsible for the production of yellow, orange, and red coloration (Duellman and Trueb 1994). The exact coloration produced by pteridines and carotenoids vary by the wavelength absorbed from these pigments (Duellman and Trueb 1994). A recent study found that the interaction between pteridine and carotenoid pigment and structural elements, such as guanine platelets found within iridophores, is associated with differences in brightness and hue (ranging in coloration from red to orange) in poison frogs of the genus Ranitomeya (Twomey et al. 2020b).
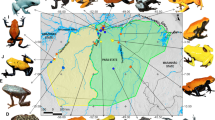
Although we have made considerable progress in identifying genes associated with color production in poison frogs, very little is known on whether these color related genes are under positive selection, with the exception of two studies (Rodríguez et al. 2020; Linderoth et al. 2023). Animal coloration is often under strong selective pressures, and thus, selection of aposematic coloration may have been episodic in nature, and may also have involved periods of stabilizing or balancing selection. Members of the genus Ranitomeya show extreme levels of color and pattern diversity both within and between species, making them an excellent system for investigating the genes underlying such diversity. In this study, we examined signatures of selection in Neotropical poison frog genus Ranitomeya, with a special emphasis on those genes putatively related to color production as coloration is strongly tied to survival in poison frogs. We investigated this by assembling transcriptomes from 9 different color morphs of poison frogs in the Ranitomeya genus that have diverse coloration (four morphs of R. imitator, two morphs of R. variabilis, two morphs of R. fantastica, one morph of R. summersi) (Fig. 1) to identify coding genes with amino acid sites under pervasive positive selection across lineages. Identification of genes responsible for coloration under positive selection may provide clues relevant to the genetic and biochemical mechanisms maintaining aposematic coloration and driving the evolution of polymorphism in poison frogs in the genus Ranitomeya.
An evolutionary tree representing the 9 color morphs of poison frogs in the Ranitomeya genus that were studied. The tree is based on Muell et al. (2022), which is a phylogenomic reconstruction of the genus Ranitomeya
Methods
Color morphs
We used data reads sets for 9 different color morphs of poison frogs in the genus Ranitomeya that exhibit diverse coloration, 8 of these data read sets were publicly available and one was published in this manuscript (Table 1). Figure 1 shows the phylogenetic tree of relationships of the poison frog morph and species in this study and is based on Muell et al. (2022), which is a phylogenomic reconstruction of the Ranitomeya genus. These include four color morphs of Ranitomeya imitator (Sauce orange-banded, Varadero red-headed, Tarapoto green-spotted, and Baja Huallaga yellow-striped morph; (Stuckert et al. 2021), two color morphs of Ranitomeya fantastica (white-banded and red-headed morph; (Stuckert et al. 2021), two color morphs of Ranitomeya variabilis (the yellow-green spotted and striped morph; (Stuckert et al. 2021), and one color morph of Ranitomeya summersi (banded morph) (published in this manuscript).
Individuals of R. imitator were laboratory reared in the United States. Breeding colonies were purchased from Understory Enterprises, LLC. Ranitomeya imitator frogs obtained from Understory Enterprises are captive bred individuals, descended directly from wild populations. As for samples of R. fantastica, R. variabilis, and R. summersi, these were captured and raised in Peru. All samples of R. imitator, R. fantastica, R. summersi, and R. variabilis were sacrificed by applying 20% benzocaine gel to their venter followed by pithing the vertebral region to ensure death. Whole skin section was then removed and stored in RNAlater (Qiagen) at 4° C for 24 h and then at -20° C until extraction. A hybrid Trizol (Ambion) protocol was used to extract RNA on all skin samples. RNA was cleaned with DNAse and RNAsin, and purified with a RNeasy spin column (Qiagen) method.
Transcriptome assembly
We downloaded publicly available skin sample reads from four unique Ranitomeya imitator color morphs (Sauce orange-banded, Varadero red-headed, Tarapoto green-spotted, and Baja Huallaga yellow-striped) (Stuckert et al. 2021). We also downloaded skin sample reads from two color morphs of R. fantastica (white-banded and red-headed) and two color morphs of R. variabilis (yellow-green spotted and striped) (Stuckert et al. 2021). In addition to the publicly available read data, we also used read data from the banded morph of Ranitomeya summersi (published in this manuscript).
We used version 2.2.2 of the Oyster River Protocol (ORP) (MacManes 2018) to generate transcriptomes from one set of paired reads per color morph. The ORP is a multi-assembler approach designed to create a high-quality transcriptome. It merges together the highest quality parts of de novo assemblies produced by Trinity (Haas et al. 2013), SPAdes (Chikhi and Medvedev 2014), and Trans-ABySS (Robertson et al. 2010) into a single high-quality assembly for each individual which we used for our analysis.
The default parameters of the ORP were used when possible. As such, the default SPAdes kmer lengths of 55 and 75 were used on all samples except R. imitator. Due to shorter R. imitator raw read lengths, we changed the SPAdes kmer lengths to 25 and 39 for these assemblies. To further improve quality, we applied a TPM filter of 1 transcript per million for each assembly. This removed lowly expressed and potentially erroneous transcripts from the data. Finally, we also included a flag that allows for strand specific assembly. We chose the RF library for strand specific assembly [https://oyster-river-protocol.readthedocs.io/en/latest/].
Detection of positive selection in protein-coding genes associated with color production
We used the pipeline FUSTr (Families Under Selection in Transcriptomes) (Cole and Brewer 2018) to identify protein-coding genes responsible for color production with amino acid sites under strong positive selection. FUSTr is a unique pipeline as it allows us to overcome the difficult task of characterizing molecular evolution in large-scale transcriptomic datasets. This method identifies and aligns homologs, and then conducts a site-specific test of positive selection. The method requires transcriptome data as input and depends on similarity of protein sequence to infer homology.
First, FUSTr identified isoforms among transcript assemblies by detecting name redundancy and then used TRANSDECODER version 3.0.1 (https://github.com/TransDecoder/TransDecoder) to predict and extract the single best open reading frame for each transcript providing nucleotide coding sequences and complementary amino acids. To ensure phylogenetic independence, the longest isoform was retained for further analysis if there were several isoforms of the same gene. FUSTr then used the program DIAMOND version 0.9.10 (Buchfink et al. 2015) to evaluate homology at the peptide level from coding sequences by implementing BLASTP with an e-value cutoff of 10− 5, and the program SiLix Version 1.2.11 (Miele et al. 2011) used transitive clustering to sort homologous coding sequences into gene families. The optimal configuration of SiLix considers sequences with a minimum identity of 35% and a minimum overlap of 90% to be similar and are added to a family (Bernardes et al. 2015). Next, FUSTr used the program MAFFT version 7.221 (Katoh and Standley 2013) to align multiple amino acid sequences of each family and used the program FastTree version 2.1.9 (Price et al. 2010) to reconstruct phylogenies of each family’s untrimmed amino acid multiple sequence alignment. Nucleotide codon sequences are used to derive trimmed multiple sequence codon alignments by reverse translating amino acid alignment. Finally, FUSTr implemented the program FUBAR (Fast Unconstrained Bayesian Approximation) (Murrell et al. 2013), which is part of the program HYPHY version 2.3.14, to conduct a site-specific test of pervasive positive selection on amino acids. The benefit of FUBAR is that it is exceptionally fast and is appropriate for analyzing large alignments. However, the limitation is that FUBAR assumes that the selection pressure for each site is constant along the entire phylogeny. We note here that sample size and speed considerations makes the application of branch and site specific methods impractical at this time. The FUBAR implements a Bayesian approach to determine nonsynonymous (dN) and synonymous substitution (dS) rates. The dN/dS ratio is a powerful measure of the strength and mode of selection acting on protein-coding sequences. Gene families with at least 15 sequences have the necessary statistical power to test for adaptive evolution and were therefore analyzed for signatures of selection (Wong et al. 2004).
We compiled all protein-coding sequences from gene families that were identified by FUSTr to be under strong positive selection and used DIAMOND version 0.9.30-3 (Buchfink et al. 2015) to perform a BLASTP search using default parameters against the uniprot database (https://www.uniprot.org/downloads). We filtered BLASTP results by keeping the best hit for each query. This allowed us to identify gene names for each protein-coding sequence from gene families that were under strong positive selection. We then identified and extracted genes associated with color pattern production (using curated lists of genes associated with coloration across vertebrates (McLean et al. 2017; Baxter et al. 2019)) from the BLASTP filtered output. Finally, we calculated the fraction of color related genes under positive selection\(\left(\frac{\text{number of color related genes under positive selection}}{\text{total number of color related genes tested}}\right)\). In addition, we calculated the fraction of color related genes under purifying selection \(\left(\frac{\text{number of color related genes under purifying selection}}{\text{total number of color related genes tested}}\right)\). We identified color related genes under purifying selection by following steps above on protein-coding sequences identified by FUSTr to have sites under purifying selection.
Gene network
We used the program GeneNetwork version 2.0 (https://www.genenetwork.nl) (Deelen et al. 2019), to investigate patterns of interactivity among all protein-coding genes associated with color production with amino acid sites under positive selection. The GeneNetwork program used gene co-regulation to predict pathway membership by using gene expression data from public 31,499 human RNA-seq samples. This program implements the GADO (GeneNetwork Assisted Diagnostic Optimization) method, which predicts phenotypic consequences of genes when mutated. It is worth noting that GeneNetwork was developed to identify networks of interactivity among genes associated with human disease. However, its results can also be used to infer connections among genes from non-model organisms. GeneNetwork implements a principal component analysis method to identify the connectivity and interaction networks between genes. The membership and biological pathways of genes influence the construction of these networks (Deelen et al. 2019). The clusters of a set of genes are those that share the same or a similar predicted pathway and function and therefore, all genes that are present in a particular cluster have similar enrichments. Nevertheless, genes from different clusters can still have strong interactions. The classification of genes with respect to color production mechanisms (e.g. melanin pathway vs. carotenoid pathway) is distinct from the biochemical pathways identified by Gene Network (based on functional pathways identified in prior research, mainly on model systems). Hence it is possible to have genes related to the same pigment class play important roles in different networks identified by Gene Network, and vice-versa (genes affecting different pigment pathways can interact in networks as identified by Gene Network).
Results
Protein-coding genes associated with color production under strong positive selection
A total of 1,294,152 transcripts from 9 samples were used as inputs into the FUSTr pipeline. FUSTr identified 1,499 families containing the minimum threshold for sufficient statistical power of ≥ 15 sequences. Of these 1,499 gene families, FUSTr identified 742 gene families with at least one amino acid site under strong positive selection (297 gene families had one amino acid site under strong positive selection, 176 gene families had two amino acid sites under strong positive selection, 89 had three amino acid sites under strong positive selection, 57 had four amino acid sites under strong positive selection, and 123 had at least five amino acid sites under strong positive selection.
We extracted and compiled all protein-coding sequences from gene families with at least one amino acid site under strong positive selection and annotated them against the uniprot database. When doing so, we found 1,227 protein-coding sequences under strong positive selection (Table S1). When cross referencing BLASTP results with a curated list based on previous research of color pattern genes known to be involved in color pattern production in other species, we found 36 genes associated with color production (Table S2). Of these, nine genes were related to melanocyte production and melanin synthesis (anxa2, ap1s1, ctbp2, dct, lef1, irf4, rab32, rab38, stx12, tcf7, tyrp1), two genes were related to iridophore development (fhl1, prps1), one gene was related to keratin metabolism (ovol1), one gene was related to pteridine synthesis (xdh), and three genes were related to carotenoid metabolism (adh1b, aldh2, rdh16) (Table 2). We found that the fraction of color related genes under positive selection\(\left(\frac{\text{number of color related genes under positive selection}}{\text{total number of color related genes tested}}\right)\) is 0.027, whereas the fraction of color related genes under purifying selection \(\left(\frac{\text{number of color related genes under purifying selection}}{\text{total number of color related genes tested}}\right)\) was 0.34.
Network analysis
We used GeneNetwork 2.0 to construct a network of genes associated with color production that were identified by FUSTr to be under positive selection. We submitted all 45 genes associated with color production under positive selection (Table S2). Of these 45 genes, 43 were recognized and used to construct a network (Fig. 2). GeneNetwork identified two separate clusters. Cluster 1 (blue) included genes associated with melanogenesis (anxa2, ap1s1, rab32), iridophore development (fhl2), and carotenoid metabolism (adh1b, aldh2). Cluster 2 (green) included genes associated with melanogenesis (ctbp2, dct, irf4, lef1, rab38, tyrp1), keratin metabolism (ovol1), pteridine synthesis (prps1, xdh), and carotenoid metabolism (rdh16). It is worth noting that this network only includes color related genes under positive selection and does not include all genes under positive selection. The reason why we only included color related genes is that this allows us to specifically visualize connections between known color related genes.
Gene Network 2.0 identified two clusters from all genes associated with color production under positive selection (Table S2). Cluster 1 (blue): includes genes associated with melanogenesis (anxa2, ap1s1, rab32), iridophore development (fhl1), and carotenoid metabolism (adh1b, aldh2). Cluster 2 (green): includes genes associated with melanogenesis (ctbp2, dct, irf4, lef1, rab38, tyrp1), iridophore development (prps1), keratin metabolism (ovol1), pteridine synthesis (xdh) and carotenoid metabolism (rdh16)
Discussion
Positive selection plays pivotal roles in adaptive evolution by promoting the emergence of novel phenotypes. Studies on comparative genetics and genomics have identified key genes that have experienced positive selection and are tied to phenotypic change in the animal kingdom (Lao et al. 2007; Williamson et al. 2007; Rubin et al. 2012; Montague et al. 2014; Rubio and Summers 2022). Although the field has made considerable progress in identifying key candidate genes associated with the production of aposematic coloration in poison frogs (Posso-Terranova and Andrés 2017; Stuckert et al. 2019, 2021; Rodríguez et al. 2020; Twomey et al. 2020a, b), very little is known about whether these color related genes are under positive selection, with the exception of two studies (Rodríguez et al. 2020; Linderoth et al. 2023).
In this study, we found 36 protein coding genes with amino acid sites under positive selection at the molecular level that were associated with color production from transcriptomes from 9 different color morphs of poison frog species in the genus Ranitomeya that have diverse forms of aposematic coloration. The protein coding genes that we found to be under strong positive selection are associated with the production of melanophores and melanin, iridophore development, pteridine synthesis, carotenoid metabolism, and keratin metabolism. Our results provide evidence of color evolution at the molecular level which may be attributed to strong selective pressures.
We also found higher rates of color related genes under purifying selection than color related genes under positive selection. Natural selection can take several different forms and intensities, however, the most prevalent form of selection is purifying selection (Nielsen et al. 2005). Purifying selection generally maintains evolutionary stasis, whereas positive selection is associated with evolutionary change and diversification across lineages. Hence, the genes found to be under positive selection may be important candidates for investigations of diversifying selection, potentially leading to polytypism or population divergence.
Melanin-related synthesis pathways
We identified several genes involved in the production of melanophores and melanin to be under positive selection (anxa2, ap1s1, ctbp2, dct, lef1, irf4, rab32, rab38, tyrp1). Genes associated with melanocyte production and melanin synthesis have a strong influence in the production of black and brown coloration. Melanic coloration is pervasive among vertebrates and a substantial set of genes influencing melanogenesis have been identified (Orteu and Jiggins 2020), mainly due to the intensive work on coat color mutation in laboratory mice (Hoekstra 2006). In vertebrates, melanic coloration is caused by the absorption of light by melanin in melanophores (Sköld et al. 2016). Melanin is synthesized from tyrosine by a key set of glycosylated enzymes (e.g., tyrosinase, tyrosinase-like protein 1 and 2), which are essential when it comes to determining the type, quantity, and quality of melanin produced (Hearing and Jiménez 1987).
The genes tyrp1 and dct, also known as tyrp2, are found within melanosomes (Marmol et al. 1996). Tyrosinase-related proteins play key roles in the melanogenesis pathway, as they interact with tyrosine and other derivatives to give rise to melanin. The tyrosinase-related protein 1 (tyrp1) gene is a critical and rate-limiting enzyme and plays an essential role in controlling what types of melanin are produced (Pawelek and Chakraborty 1998). In addition, tyrp1 has been found to be associated with melanoma maintenance and influences melanocyte proliferation and melanocyte cell death (Johnson and Jackson 1992; Kobayashi et al. 1998; Hirobe and Abe 1999). The tyrp1 gene was differentially expressed between developmental stages in R. imitator and R. variabilis (Stuckert et al. 2021) of D. auratus (Stuckert et al. 2019), and between color morphs of O. pumilio (Rodríguez et al. 2020). In the polymorphic tawny dragon lizard, Ctenophorus decresii, tyrp1 was found to be upregulated in grey colored throats when compared to orange, yellow, and blue throats (McLean et al. 2017). The gene Rab38, which has the highest network connectivity in cluster 2, plays vital roles in melanosomal protein trafficking and melanocyte pigmentation. Findings from Wasmeier et al. (2006) suggest that rab38 is not necessary for early stage melanosome formation, however, the absence of this gene results in a dramatic loss of pigmentation. This finding supports the notion that rab38 is critical in the trafficking of tyrp1 and tyrosinase (Watabe et al. 2004; Hearing 2005).
Iridophore/guanine platelet pathways
We identified genes involved in iridophore development (fhl1, prps1) to be under positive selection. Iridophores are associated with the production of white, green, and blue coloration; ultimately determined by the amount of light reflected from their internal structural elements (Bagnara et al. 2007). Iridophores contain reflective stacked guanine platelets that are nested in cytoplasm (Johnsen 2012). By using reflectance spectrometry, pigmentation analysis, electron microscopy, and color modeling, Twomey et al. (2020b) found that in poison frogs, the thickness of these structural elements (in combination with pigmentation) determines skin hue. For example, orange coloration was produced when guanine platelets are thick in size reflecting long wavelengths, whereas blue coloration is produced when platelets are thin in size, reflecting short wavelengths. This suggests that variation in the thickness of guanine platelets plays a vital role in controlling overall coloration in poison frogs. The mechanisms underlying the formation of guanine platelets in iridophores remains unclear however, it has been suggested that Rab GTPases may play key roles in this context (Higdon et al. 2013). Studies suggest that epidermis-structuring genes may also be playing vital roles in the formation of structural colors, however, more evidence is necessary to pinpoint the exact role of these genes (McGowan et al. 2006; Stuckert et al. 2019; Burgon et al. 2020). The production keratin and organization of the dermis can influence structural colors in a similar way that guanine platelets influence structural colors. The gene fhl1 (four and a half LIM domains protein 1), which was placed in cluster 1 (blue) of our gene network, is expressed in striated muscle (Chu et al. 2000; Tanahashi and Tabira 2000). We also identified the ovol1 (ovo like transcriptional repressor 1) gene, which is involved in epidermal keratinization (Tsuji et al. 2018), to be under positive selection. Nair et al. (2006) demonstrated that ovol1-deficient exhibited hyperproliferative skin disorders, demonstrating the crucial role ovol1 plays in the formation of epidermal tissue. It is possible that the ovol1 gene is playing vital roles in the formation of structural colors via the production of epidermal tissue in poison frogs.
Xanthophores: pteridine pathways and carotenoid metabolism
We identified genes involved in the production of pteridine synthesis (xdh) and carotenoid metabolism (adh1b, aldh2, rdh16) to be under positive selection. Pteridine and carotenoids are both pigments contained in xanthophores (Duellman and Trueb 1994). Xanthophores are associated with the production of yellow, orange, and red coloration (Obika and Bagnara 1964; Grether et al. 2001; Mcgraw et al. 2006; Croucher et al. 2013; McLean et al. 2017). The exact coloration derived varies by the wavelength absorbed from both pteridine and carotenoid pigments (Duellman and Trueb 1994). Pteridines are purine-based pigments which are synthesized internally by many animals (Stackhouse 1966). Pteridine pigments are derived as they are synthesized during the production of purines (Steffen and McGraw 2009). The production of guanine is a fundamental physiological process, and therefore, pteridines are continuously replenished. However, not all pigments are produced de novo by animals. For instance, animals are incapable of synthesizing carotenoids de novo, and must be acquired through the diet (Goodwin 1992). In animals, dietary provitamin A carotenoids (e.g. beta-carotene) are vital as they promote growth and overall fitness of organisms (Hill et al. 2006). In addition, numerous studies indicate that provitamin A promotes the production and maintenance of bright coloration (e.g., red, orange) (Ahi et al. 2020; Twomey et al. 2020a).
The prps1 gene (phosphoribosyl pyrophosphate synthetase 1) catalyzes the synthesis of phosphoribosyl pyrophosphate, which is essential for the de novo synthesis of purine, pyrimidine, and pyridine nucleotides (Kornberg et al. 1955; Lieberman et al. 1955; Hartman and Buchanan 1958; Preiss and Handler 1958). The gene xdh (xanthine hydrogenase) is well known to play crucial roles in the synthesis of pteridines, which are pigments deposited in xanthophores that play crucial roles in yellow, orange and red coloration (Epperlein and Löfberg 1990; Bagnara and Matsumoto 2006). For instance, xdh plays vital roles in producing red drosopterin that are placed in the eyes of Drosophila flies (Reaume et al. 1991). In whole skin tissue, xdh was found to be differentially expressed between color morphs of O. pumilio (Rodríguez et al. 2020), between color morphs of D. auratus (Stuckert et al. 2019), and between larval developmental stages of R. variabilis and R. imitator (Stuckert et al. 2021).
The genes adh1b (alcohol dehydrogenase 1B) and aldh2 (aldehyde dehydrogenase 2) genes both play vital roles in the synthesis of retinoic acid. The first step of retinoic acid synthesis is the oxidation of retinol to retinaldehyde, which are catalyzed by genes in the alcohol dehydrogenase (ADH) and retinol dehydrogenase (RDH) family, and the second step is the oxidation of retinaldehyde to produce retinoic acid, which is managed by genes in the aldehyde dehydrogenase (ALDH) family (Duester 2000; Molotkov et al. 2002; Molotkova et al. 2007). Retinoic acid is vital as it controls growth and development by metabolizing retinol.
Conclusion
We identified protein-coding genes responsible for color production that are under pervasive positive selection using transcriptome data from a set of poison frog species in the genus Ranitomeya. All poison frog species and morphs used in this study exhibit bright conspicuous coloration accompanied with contrasting patterns. Members of the family Dendrobatidae rely on bright aposematic coloration to communicate to potential predators that they are poisonous and unpalatable. Numerous studies support the notion that bright conspicuous coloration is strongly tied to survival in poison frogs and therefore, the genetic mechanisms responsible for production of aposematic coloration are likely to have come under positive selection. We implemented the pipeline FUSTr and identified multiple genes under strong positive selection related to color production. This included genes that play key roles in melanocyte production and melanin synthesis, iridophore development, keratin metabolism, pteridine synthesis, and carotenoid metabolism. Several genes found to be under positive selection in our study (e.g., xdh, tyrp1, dct, irf4) have been found to be differentially expressed in other poison frog studies; between morphs of Dendrobates auratus (Stuckert et al. 2019), and between larval developmental stages and among color morphs of R. imitator, R. variabilis, and R. fantastica (Stuckert et al. 2021). Ultimately, understanding the roles of changes in gene regulation and changes in coding sequence in the evolution of diversity in coloration in the poison frogs (between individuals, populations and species) will require exhaustive elucidation of gene regulatory mechanisms, patterns of differential expression, and the identification of coding sequence differences that are under positive selection. In this paper, we have begun to elucidate key color-related genes that have been under selection in this lineage. This data will be crucial as we work towards understanding the combined effects of coding sequence changes and changes in gene regulation (and their interaction) in driving the remarkable diversity in coloration in this group.
Data availability
Raw read data for Ranitomeya summersi are archived with the National Center for Biotechnology Information (Accession number: SAMN39933221). All transcriptomes used in this manuscript are available on Zenodo (https://zenodo.org/records/10893167).
References
Ahi EP, Lecaudey LA, Ziegelbecker A et al (2020) Comparative transcriptomics reveals candidate carotenoid color genes in an east African cichlid fish. BMC Genomics 21:54
Bagnara JT, Matsumoto J (2006) Comparative anatomy and physiology of pigment cells in nonmammalian tissues. The pigmentary system: physiology and pathophysiology 11–59
Bagnara JT, Fernandez PJ, Fujii R (2007) On the blue coloration of vertebrates. Pigment Cell Res 20:14–26
Baxter LL, Watkins-Chow DE, Pavan WJ, Loftus SK (2019) A curated gene list for expanding the horizons of pigmentation biology. Pigment Cell Melanoma Res 32:348–358
Bernardes JS, Vieira FRJ, Costa LMM, Zaverucha G (2015) Evaluation and improvements of clustering algorithms for detecting remote homologous protein families. BMC Bioinformatics 16:34
Briolat ES, Zagrobelny M, Olsen CE et al (2018) Sex differences but no evidence of quantitative honesty in the warning signals of six-spot burnet moths (Zygaena filipendulae L). https://doi.org/10.1111/evo.13505. Evolution
Briolat ES, Burdfield-Steel ER, Paul SC et al (2019) Diversity in warning coloration: selective paradox or the norm? Biol Rev Camb Philos Soc 94:388–414
Brown JL (2013) The evolution of parental care, aposematism and color diversity in neotropical poison frogs. Evol Ecol 27:825–829
Brown JL, Twomey E, Amézquita A et al (2011) A taxonomic revision of the neotropical poison frog genus Ranitomeya (Amphibia: Dendrobatidae). Zootaxa 3083:1
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Burgon JD, Vieites DR, Jacobs A et al (2020) Functional colour genes and signals of selection in colour-polymorphic salamanders. Mol Ecol 29:1284–1299
Caro T, Sherratt TN, Stevens M (2016) The ecology of multiple colour defences. Evol Ecol 30:797–809
Chai P (1986) Field observations and feeding experiments on the responses of rufous-tailed jacamars (Galbula ruficauda) to free-flying butterflies in a tropical rainforest. Biol J Linn Soc Lond 29:161–189
Chikhi R, Medvedev P (2014) Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31–37
Chu PH, Ruiz-Lozano P, Zhou Q et al (2000) Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech Dev 95:259–265
Cole TJ, Brewer MS (2018) FUSTr: a tool to find gene families under selection in transcriptomes. PeerJ 6:e4234
Croucher PJP, Brewer MS, Winchell CJ et al (2013) De novo characterization of the gene-rich transcriptomes of two color-polymorphic spiders, Theridion grallator and T. californicum (Araneae: Theridiidae), with special reference to pigment genes. BMC Genomics 14:862
Cummings ME, Crothers LR (2013) Interacting selection diversifies warning signals in a polytypic frog: an examination with the strawberry poison frog. Evol Ecol 27:693–710
Cuthill IC, Allen WL, Arbuckle K et al (2017) The biology of color. Sci 357. https://doi.org/10.1126/science.aan0221
Deelen P, van Dam S, Herkert JC et al (2019) Improving the diagnostic yield of exome- sequencing by predicting gene-phenotype associations using large-scale gene expression analysis. Nat Commun 10:2837
Duellman WE, Trueb L (1994) Biology of amphibians. JHU
Duester G (2000) Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem 267:4315–4324
Epperlein HH, Löfberg J (1990) The development of the larval pigment patterns in Triturus alpestris and Ambystoma mexicanum. Adv Anat Embryol Cell Biol 118:1–99
Finkbeiner SD, Briscoe AD, Reed RD (2014) Warning signals are seductive: relative contributions of color and pattern to predator avoidance and mate attraction in Heliconius butterflies. Evolution 68:3410–3420
Fox DL (1936) Structural and chemical aspects of animal coloration. Am Nat 70:477–493
Goodwin TW (1992) [15] distribution of carotenoids. Methods in Enzymology. Academic, pp 167–172
Gray SM, McKinnon JS (2007) Linking color polymorphism maintenance and speciation. Trends Ecol Evol 22:71–79
Grether GF, Millie DF, Bryant MJ et al (2001) Rain forest canopy cover, resource availability, and life history evolution in guppies. Ecology 82:1546–1559
Haas BJ, Papanicolaou A, Yassour M et al (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Hartman SC, Buchanan JM (1958) Biosynthesis of the purines. XXI. 5-Phosphoribosylpyrophosphate amidotransferase. J Biol Chem 233:451–455
Harvey PH, Bull JJ, Pemberton M, Paxton RJ (1982) The evolution of Aposematic Coloration in Distasteful Prey: A Family Model. Am Nat 119:710–719
Hearing VJ (2005) Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci 37:3–14
Hearing VJ, Jiménez M (1987) Mammalian tyrosinase–the critical regulatory control point in melanocyte pigmentation. Int J Biochem 19:1141–1147
Hebets EA, Papaj DR (2005) Complex signal function: developing a framework of testable hypotheses. Behav Ecol Sociobiol 57:197–214
Higdon CW, Mitra RD, Johnson SL (2013) Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS ONE 8:e67801
Hill GE, Hill GE, McGraw KJ, McGraw, KJ (2006) Bird coloration: mechanisms and measurements. Harvard University Press
Hirobe T, Abe H (1999) Genetic and epigenetic control of the proliferation and differentiation of mouse epidermal melanocytes in culture. Pigment Cell Res 12:147–163
Hoekstra HE (2006) Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97:222–234
Johnsen S (2012) The Optics of Life: a biologist’s guide to light in Nature. Princeton University Press
Johnson R, Jackson IJ (1992) Light is a dominant mouse mutation resulting in premature cell death. Nat Genet 1:226–229
Joron M, Mallet JL (1998) Diversity in mimicry: paradox or paradigm? Trends Ecol Evol 13:461–466
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kobayashi T, Imokawa G, Bennett DC, Hearing VJ (1998) Tyrosinase stabilization by Tyrp1 (the brown locus protein). J Biol Chem 273:31801–31805
Kokko H, Brooks R, McNamara JM, Houston AI (2002) The sexual selection continuum. Proc Biol Sci 269:1331–1340
Kornberg A, Lieberman I, Simms ES (1955) Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J Biol Chem 215:389–402
Lao O, de Gruijter JM, van Duijn K et al (2007) Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet 71:354–369
Lieberman I, Kornberg A, Simms ES (1955) Enzymatic synthesis of pyrimidine nucleotides; orotidine-5’-phosphate and uridine-5’-phosphate. J Biol Chem 215:403–451
Linderoth T, Aguilar-Gómez D, White E et al (2023) Genetic basis of aposematic coloration in a mimetic radiation of poison frogs. bioRxiv 2023.04.20.537757
Longley WH (1917) Studies upon the Biological significance of animal coloration. II. A revised Working Hypothesis of Mimicry. Am Nat 51:257–285
Losos JB (1985) An experimental demonstration of the species-Recognition Role of Anolis Dewlap Color. Copeia 1985:905–910
Maan ME, Cummings ME (2009) Sexual dimorphism and directional sexual selection on aposematic signals in a poison frog. Proc Natl Acad Sci U S A 106:19072–19077
MacManes MD (2018) The Oyster River Protocol: a multi-assembler and kmer approach for de novo transcriptome assembly. PeerJ 6:e5428
Mallet J, Joron M (1999) Evolution of diversity in warning color and mimicry: polymorphisms, shifting balance, and speciation. Annu Rev Ecol Syst 30:201–233
Marmol Vdel, del Marmol V, Beermann F (1996) Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett 381:165–168
McGowan KA, Aradhya S, Fuchs H et al (2006) A mouse keratin 1 mutation causes dark skin and epidermolytic hyperkeratosis. J Invest Dermatol 126:1013–1016
Mcgraw KJ, Nolan PM, Crino OL (2006) Carotenoid accumulation strategies for becoming a colourful House Finch: analyses of plasma and liver pigments in wild moulting birds. Funct Ecol 20:678–688
McLean CA, Lutz A, Rankin KJ et al (2017) Revealing the Biochemical and Genetic Basis of Color Variation in a polymorphic lizard. Mol Biol Evol 34:1924–1935
Miele V, Penel S, Duret L (2011) Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12:116
Molotkov A, Deltour L, Foglio MH et al (2002) Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J Biol Chem 277:13804–13811
Molotkova N, Molotkov A, Duester G (2007) Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev Biol 303:601–610
Montague MJ, Li G, Gandolfi B et al (2014) Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A 111:17230–17235
Muell MR, Chávez G, Prates I et al (2022) Phylogenomic analysis of evolutionary relationships in Ranitomeya poison frogs (Family Dendrobatidae) using ultraconserved elements. Mol Phylogenet Evol 168:107389
Müller F (1879) Ituna and Thyridia: a remarkable case of mimicry in butterflies. Trans Entomol Soc Lond 1879:20–29
Murrell B, Moola S, Mabona A et al (2013) FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205
Nair M, Teng A, Bilanchone V et al (2006) Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol 173:253–264
Nielsen R, Bustamante C, Clark AG et al (2005) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 3:e170
Obika M, Bagnara JT (1964) Pteridines as pigments in amphibians. Science 143:485–487
Orteu A, Jiggins CD (2020) The genomics of coloration provides insights into adaptive evolution. Nat Rev Genet 21:461–475
Pawelek JM, Chakraborty AK (1998) The enzymology of melanogenesis. The pigmentary system: physiology and pathophysiology 391–400
Posso-Terranova A, Andrés JÁ (2017) Diversification and convergence of aposematic phenotypes: truncated receptors and cellular arrangements mediate rapid evolution of coloration in harlequin poison frogs. Evolution 71:2677–2692
Preiss J, Handler P (1958) Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem 233:493–500
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490
Reaume AG, Knecht DA, Chovnick A (1991) The rosy locus in Drosophila melanogaster: xanthine dehydrogenase and eye pigments. Genetics 129:1099–1109
Reynolds RG, Fitzpatrick BM (2007) Assortative mating in poison-dart frogs based on an ecologically important trait. Evolution 61:2253–2259
Robertson G, Schein J, Chiu R et al (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7:909–912
Rodríguez A, Mundy NI, Ibáñez R, Pröhl H (2020) Being red, blue and green: the genetic basis of coloration differences in the strawberry poison frog (Oophaga pumilio). BMC Genomics 21:301
Rubin C-J, Megens H-J, Martinez Barrio A et al (2012) Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci U S A 109:19529–19536
Rubio AO, Summers K (2022) Neural crest cell genes and the domestication syndrome: a comparative analysis of selection. PLoS ONE 17:e0263830
Rudh A, Qvarnström A (2013) Adaptive colouration in amphibians. Semin Cell Dev Biol 24:553–561
Ruxton GD, Sherratt TN, Speed MP (2004) Avoiding attack: the Evolutionary Ecology of Crypsis, warning signals and mimicry. Oxford University Press
Saenko SV, Teyssier J, van der Marel D, Milinkovitch MC (2013) Precise colocalization of interacting structural and pigmentary elements generates extensive color pattern variation in Phelsuma lizards. BMC Biol 11:105
Saporito RA, Zuercher R, Roberts M et al (2007) Experimental Evidence for Aposematism in the Dendrobatid Poison Frog Oophaga pumilio. Copeia 2007:1006–1011
Sköld HN, Aspengren S, Cheney KL, Wallin M (2016) Fish Chromatophores–from Molecular motors to Animal Behavior. Int Rev Cell Mol Biol 321:171–219
Stackhouse HL (1966) Some aspects of pteridine biosynthesis in amphibians. Comp Biochem Physiol 17:219–235
Steffen JE, McGraw KJ (2009) How dewlap color reflects its carotenoid and pterin content in male and female brown anoles (Norops sagrei). Comp Biochem Physiol B Biochem Mol Biol 154:334–340
Stuckert AMM, Moore E, Coyle KP et al (2019) Variation in pigmentation gene expression is associated with distinct aposematic color morphs in the poison frog, dendrobates auratus. BMC Evol Biol 19:1–15
Stuckert AMM, Chouteau M, McClure M et al (2021) The genomics of mimicry: gene expression throughout development provides insights into convergent and divergent phenotypes in a Müllerian mimicry system. Mol Ecol 30:4039–4061
Stuckert AMM, Freeborn L, Howell KA et al (2023) Transcriptomic analyses during development reveal mechanisms of integument structuring and color production. Evol Ecol 1–22
Summers K, Symula R, Clough M, Cronin T (1999) Visual mate choice in poison frogs. Proc Biol Sci 266:2141–2145
Tanahashi H, Tabira T (2000) Alzheimer’s disease-associated presenilin 2 interacts with DRAL, an LIM-domain protein. Hum Mol Genet 9:2281–2289
Tsuji G, Ito T, Chiba T et al (2018) The role of the OVOL1-OVOL2 axis in normal and diseased human skin. J Dermatol Sci 90:227–231
Twomey E, Vestergaard JS, Venegas PJ, Summers K (2016) Mimetic divergence and the Speciation Continuum in the mimic poison Frog Ranitomeya imitator. Am Nat 187:205–224
Twomey E, Johnson JD, Castroviejo-Fisher S, Van Bocxlaer I (2020a) A ketocarotenoid-based colour polymorphism in the Sira poison frog Ranitomeya sirensis indicates novel gene interactions underlying aposematic signal variation. Mol Ecol 29:2004–2015
Twomey E, Kain M, Claeys M et al (2020b) Mechanisms for Color Convergence in a Mimetic Radiation of poison frogs. Am Nat 195:E132–E149
Wasmeier C, Romao M, Plowright L et al (2006) Rab38 and Rab32 control post-golgi trafficking of melanogenic enzymes. J Cell Biol 175:271–281
Watabe H, Valencia JC, Yasumoto K-I et al (2004) Regulation of tyrosinase processing and trafficking by organellar pH and by proteasome activity. J Biol Chem 279:7971–7981
Williamson SH, Hubisz MJ, Clark AG et al (2007) Localizing recent adaptive evolution in the human genome. PLoS Genet 3:e90
Wong WSW, Yang Z, Goldman N, Nielsen R (2004) Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051
Yang Y, Richards-Zawacki CL, Devar A, Dugas MB (2016) Poison frog color morphs express assortative mate preferences in allopatry but not sympatry. Evolution 70:2778–2788
Ziegler I (2003) The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res 16:172–182
Acknowledgements
We thank Wilson X. Guillory for poison frog silhouettes drawings that were used in this manuscript. We are also grateful to the anonymous reviewers for their insightful comments and suggestions that improved this manuscript.
Funding
Open access funding provided by the Carolinas Consortium.
Author information
Authors and Affiliations
Contributions
Designed research: AOR, KSTranscriptome assembly: AMMS, TMLAnalyzed FUSTr results: AOR, TJCContributed to writing: AOR, AMMS, TML, TJC, KSAll authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Animal care and use for this research was approved by East Carolina University’s IACUC (AUP 281b).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rubio, A.O., Stuckert, A.M.M., LaPolice, T.M. et al. Under pressure: evidence for selection on color-related genes in poison frogs of the genus Ranitomeya. Evol Ecol (2024). https://doi.org/10.1007/s10682-024-10297-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10682-024-10297-1