Abstract
Sorghum (Sorghum bicolor L. Moench) has become an increasingly valuable crop for food, feed, and especially bioenergy feedstock production, which makes the crop extremely attractive for studying genomics and genetic diversity. Molecular markers and genomics play essential roles in sorghum breeding. The rapid development of next-generation sequencing technology has facilitated the identification of genome-wide insertion-deletion (InDel) polymorphisms, enabling the efficient construction of InDel markers that are suitable for user-friendly PCR. This study was conducted with the objective of discovering and developing InDel markers using double digest restriction site-associated DNA sequencing (ddRAD-Seq) data. A total of 19,226 InDels distributed across 10 chromosomes in the sorghum genome was identified. Of those, deletions constituted 65.7% while the remain was insertions. A comprehensive analysis of all the chromosomes revealed a total of 80 InDel sites with a minimum length of 10 bp. For a good conversion of the InDel regions to beneficial molecular markers, specific primers were designed for the amplification of 47 InDel regions that were selected for further investigation. A diverse panel of sorghum consisting of 16 accessions served a source for the developed InDel markers validation. Of the 47 InDel markers, 14 were tested across 16 sorghum accessions and were demonstrated their helpfulness for marker-assisted selection in sorghum. The polymorphic information content (PIC) values of the 16 markers varied between 0.11 and 0.38, with an average of 0.28. The findings of this study indicated that the identification of InDels and the development of molecular markers for sorghum were accomplished using the ddRAD-Seq data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorghum [Sorghum bicolor (L.) Moench] is a member of the Poaceae family, originated in northeastern Africa (Doggett 1988; Kimber 2000). It is widely distributed throughout the world and has various uses, such as in staple foods, animal feed, and as a resource of raw material for bioethanol production (Mathur et al. 2017). Sorghum exhibits wide ecological adaptability, thrives in various climates ranging from arid regions with minimal rainfall to high rainfall and humid areas, as well as temperate regions worldwide (Techale et al. 2022). Moreover, sorghum displays salt tolerance and water-logging resistance (Almodares and Hadi 2009). Compared to prominent cereal crops like wheat, maize, and rice, it exhibits lower input requirements and higher yields (Mathur et al. 2017). With all these advantages, sorghum is considered a “failsafe” crop in agricultural ecosystems (Paterson 2008).
Assessment of genetic diversity in a crop is of high importance in the fields of germplasm management, application, and genotype selection for the purpose of enhancing agricultural development. Genetic diversity can be investigated through molecular markers that can provide rapid and accurate characterization that is unaffected by environmental changes (Jones et al. 2009; Mamo et al. 2023). Various molecular markers, such as Amplified Fragment Length Polymorphism (AFLP) (Gerrano et al. 2014; Medraoui et al. 2024), Random Amplified Polymorphic DNA (RAPD) (Raza et al. 2019; Ruiz-Chutan et al. 2019), Restriction Fragment Length Polymorphism (RFLP) (Cui et al. 1995; Ahnert et al. 1996), Diversity Arrays Technology (DArT) (Mace et al. 2020), Simple Sequence Repeats (SSR) (Zheng et al. 2021; Kedir et al. 2024), and Single Nucleotide Polymorphism (SNP) (Enyew et al. 2022; Yahaya et al. 2023) have been employed in the identification of genetic diversity in sorghum breeding.
Insertion/deletion polymorphisms are a class of markers that arise via many mechanisms, including the insertion of transposable elements, slippage during simple sequence replication, or uneven crossover events (Britten et al. 2003). In comparison to other types of molecular markers, InDel markers have several advantages including abundant presence within the genome, accurate identification capability, reduced demands for high-quality and large quantities of DNA samples for testing, and a simplified experimental process that requires only commonly available equipment (Gao et al. 2012). The number of studies in which InDel markers have been developed and validated has increased significantly because of all these advantages. For example, a large number of InDels was identified in the Mediterranean sesame collection using a ddRAD-Seq protocol developed by Kizil et al. (2020). Out of these, 16 InDels were evaluated across 32 sesame accessions. Liu et al. (2015) developed 48 InDel markers by using conserved sequences derived from functional genes in peanut. These markers were validated in comprehensive panel consisting of 118 different accessions. Ren et al. (2023) identified 195 InDel markers. To further validate, 27 of them were used for 47 double flower cultivars and 22 single flower cultivars of Prunus mume. The process of discovering Indel markers is comparatively sluggish in sorghum when compared to other crops, and there are currently limited studies developing and validating InDel markers in sorghum. Most of InDels were identified in two Sudan grass varieties (Sa and S722) using a RADSeq method (Wang et al. 2022). Out of these, 100 InDels were evaluated across between sorghum Tx623B and Sa and between Tx623B and S722. Choe et al. (2023), two InDel markers with sizes greater than 20 bp were developed for the purpose of discriminating between different cytotypes and validated a total of 1104 plants from six sorghum cultivars to determine variant cytotypes.
Significant advancements have been achieved in sorghum breeding because it has garnered considerable interest in recent years as a potential “star” crop. The Sorghum genome is fully sequenced. This information has contributed to the development of molecular markers such as InDels, which have many advantages, and the assessment of the level of genetic variation which serve as the basis for genetic material selection in plant breeding. The objectives of this study were to: (1) detect InDels using the ddRAD-Seq method and generate PCR-based InDel markers; (2) validate InDel markers in a diverse sorghum panel; and (3) examine genetic diversity and relationships in the sorghum panel.
Materials and methods
Plant materials and DNA extraction
Twenty-seven sorghum accessions (two sweet and one grain sorghum cultivars) were used as genetic materials in this study (Table S1). The seeds of each genotype were sown in pots, and young leaves were collected. The DNAs were extracted using the CTAB method with minor modifications (Doyle and Doyle 1990). The final concentration of DNA was normalized to 100 ng/μL using lambda DNA as a reference.
Library construction and sequencing
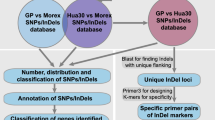
A modified version of the ddRAD-seq method (Peterson et al. 2012) was followed for genomic library construction with eleven accessions. Briefly, the genomic DNA was subjected to restriction enzymes; VspI (ATTAAT) and MspI (CCGG) and then Illumina paired-end adaptors (P1 and P2, P1 adapter's 3' end was modified to match the overhanging VspI restriction cut site) were ligated to the fragmented DNA using T4 DNA ligase buffer (Peterson et al. 2012). PCR was used to amplify with genotype-specific indexed PCR primers. The ligated DNA products were visualized on 2% agarose gel and selected based on the average size of 400–500 bp DNA fragments. The resulting ddRAD-seq genomic library was sequenced on an Illumina Hi-Seq platform via 150 bp paired-end sequencing.
InDel calling
InDel calling was performed in the following steps:
-
1.
The raw reads were demultiplexed with Je V1.2 (Girardot et al. 2016),
-
2.
Trimming of adaptors and filtering was conducted with fastp (v0.20.1) (Chen et al. 2018) (Phred quality score less than 15 out of 40),
-
3.
The filtered reads were aligned with sorghum reference genome (McCormick et al. 2018) using Bowtie2 (v2.3.4.3) (Langmead and Salzberg 2012),
-
4.
The created BAM files analyzed, and variants were called with freebayes (v13.1) (Garrison and Marth 2012) (simple diploid calling and filtering, and coverage values of 20x),
-
5.
VCF files were filtered with (SNPs were discarded) VCFfilter (Galaxy Version 1.0.0),
-
6.
All.vcf files were combined with (individual.vcf files included insertions and deletions; with VCF genotypes (Galaxy Version 1.0.0).
-
7.
The merged variant file underwent processing in Microsoft Excel to remove duplicate areas and arrange the InDels based on their sizes.
-
8.
The InDel regions with a minimum length of 10 base pairs, were evaluated using the Integrated Genome Browser V9.1.4 (IGB) (Freese et al. 2016). This analysis was conducted for each BAM file in conjunction with the sorghum reference genome.
Primer design and PCR analysis
To generate genome-wide InDel markers, flanking sequences of selected insertion-deletion regions were used. Primers were designed with Primer3Plus (Untergasser et al. 2007) with the following parameters: predicted PCR products ranged from 100 to 900 bp; the length of primers was limited to 18 bp; the annealing temperature was restricted to 50–58 °C. The primer pairs were subsequently checked with Integrated Genome Browser V9.1.4 (Freese and Norris 2016), for possible sequence duplications in the genome. All markers were given a name with the format SG-D(I)-X-XXX, where "SG" defines sorghum, "D" and "I" indicate deletion and insertion, "X" is the chromosome number, and "XXX" denotes the beginning of the chromosomal position. The InDel annotation was derived from the reference genome sequence of sorghum using Generic Feature Format 3 (Sorghum_bicolor_NCBIv3.54.gff3).
The PCR analyses were performed in 15 μl of reaction mixture containing 1 μl DNAs (40 ng), 1.5 μl 10 × PCR buffer, 2.5 mM MgCl2, 1.5 μl dNTP mix (4 mM), 1 μl each of forward and reverse primers (10 μM), 0.2 μl of Taq DNA polymerase (5 U/μl), and amount of Milli-Q water. The thermal cycles include 94 °C for 5 min, 30 cycles of 94 °C for 59 s, 50–58 °C for 59 s and 72 °C for 1 min, with an extension 72 °C for 10 min. The PCR products were analyzed by 3% agarose gel electrophoresis and visualized by UV light.
Validation of developed InDel markers
A total of 16 accessions with diverse genetic backgrounds were screened with the developed 14 InDel markers. The predicted PCR bands were visualized with a Fragment AnalyzerTM (Advanced Analytical Technologies GmbH, Heidelberg, Germany) with the use of DNF-900-K0500 reagent kit for the qualitative analysis of DNA fragments. The solutions, buffers, and gels were used in accordance with the instructions provided by the manufacturers. The bands were visualized in virtual gel which show size of bands ranging from 1 base pair (bp) as the lowest and 500 bp as the highest. Additionally, calibration was performed using a DNA ladder within the 1 to 500 bp range. The obtained data was evaluated using PROSize 2.0 (Advanced Analytical Technologies, AMES, IA, USA).
Genetic diversity analysis
Population genetic parameters, including number of alleles (Na), effective number of alleles (Ne), Shannon diversity index (I), expected heterozygosity (He), and principal coordinate analysis (PCoA) were calculated using GenAlex V6.5 (Peakall et al. 2012). The polymorphism measurement was performed using the Excel Microsatellite Toolkit (Park 2002).
Results
After quality filtering, a total of 33.7 M reads was obtained by paired-end sequencing. The mean number of reads was 3.06 M and the guanine-cytosine (GC) content was 40.32% among the sequenced genotypes. The highest and lowest reads were acquired by BSS47 genotype and Ogretmenoglu variety, respectively. The mean of the ddRAD reads mapped to the Sorghum bicolor reference genome was 81.89%.
We identified 19,226 InDel sites following to the bioinformatic pipeline. The deletions were composed of 65.7% of the total InDel positions, and their sizes varied from 1 to 14 bp, and 90.8% of all deletions had a size length of 1 to 2 bp. Among the insertions, 95.4% were less than 5 bp length, 4.5% were between 5 and 10 bp, and 0.3% were more than 10 bp (Table 1).
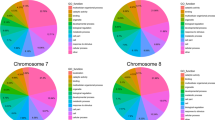
A total of 4814 InDel regions were identified, each with a size of ≥ 2 bp, distributed throughout the genome. The number of markers per chromosome ranged from 347 to 730 (Table 2).The identified deletion markers were 2903which were higher than the number of insertion markers which was 1911. Furthermore, variations in the distribution of insertions and deletions were observed across different chromosomes. Chromosome 1 had the highest number of deletions with 483, while chromosome 8 had the lowest number of insertions with 150. The frequency of InDels (size ≥ 2 bp) varied among the chromosomes, ranging from 5.53 to 9.02 InDels/Mb. The highest frequency of deletions and insertions was observed on chromosomes 1 and 2, while chromosome 8 showed the lowest frequency (Table 2).
A total of 80 InDel sites (≥ 10 bp) was detected in the sorghum genome (Table 3). Chromosome 10 exhibited the highest number of deletions (9), while chromosome 2 displayed the greatest number of insertions (10). There was no deletion and insertion of ≥ 10 bp in chromosomes 1 and 3, respectively (Table 3). Chromosomes 8 and 9 each exhibited only one deletion with a length of ≥ 10 bp. The chromosome 8 exhibited the longest insertion, spanning 14 bp, at a physical location of 54,175,764 (Table S2). On the other hand, the longest deletion, spanning 14 bpeach, was shown in chromosome 4 (physical position: 10,180,777), chromosome 5 (physical position: 10,427,959), and chromosome 6 (physical position: 47,562,364) (Table S3). The Integrated Genome Browser (IGB) also visualized the genomic positions of selected insertions and deletions (Figs. S1 and S2).
The primer pairs were designed with the use of 47 insertion-deletions that were identified across all the 10 chromosomes that have a length of 10 bp and/or greater. The highest number of markers was designed in chromosome 2 (9 markers), followed by chromosome 10, with 8 markers in total, including 7 deletion and 1 insertion marker. The primers used in this study successfully generated clean amplicons, as shown in Fig. S3. The analysis of insertion-deletion genomic positions revealed that the majority of those (74.6%) was located in intergenic regions. A smaller proportion was found in messenger RNA regions (mRNA) (12.7%), introns (6.4%), exons (4.2%), and coding sequences (CDS) (2.1%) (Table S4).
We tested 14 InDel markers (lengths ≥ 10 bp) with genetic diversity analysis using 16 sorghum accessions that originated from nine countries. The PCR products varied between 100 and 600 bp, which were visualized on a Fragment Analyzer® (Fig. S4). The InDel markers exhibited predicted polymorphisms among the accessions and were submitted in Table 4. The expected heterozygosity was determined to range from 0.000 to 0.135. The loci SG-D-2-581 and SG-I-6-563 exhibited the greatest expected heterozygosity value. The average expected heterozygosity across all loci was determined to be 0.072. The average Shannon diversity index (I) was determined to be 0.101. The range of polymorphic information content (PIC) values for these 14 markers varied from 0.110 to 0.380, with an average value of 0.282. The results of Principal Coordinate Analysis (PCoA) revealed that the first and second coordinates accounted for 26.50% and 17.04% of the overall variance, respectively. The sorghum accessions were further categorized based on the inclusion of accessions originating from various continents; they were divided into three groups in the PCoA graphic. The first group (I) included seven genotypes (BSS206, BSS224, BSS241, BSS244, BSS283, and BSS293) from three continents. The second (II) and third (III) groups contained four (BSS249, BSS269, BSS270, and BSS295) from four continents and five (BSS203, BSS210, BSS221, BSS232, and BSS292) from one continent, respectively (Fig. 1).
Discussion
With the advent of next-generation sequencing (NGS) technologies, different types of genome reduction methods were developed, which has provided effective opportunities for generating data for marker development. ddRAD-Seq is one of the reduced-representation sequencing approaches to identify variations in the studied genome. The method provided lots of data to identify the genetic diversity and population structure of the studied sorghum panel. In this study, we identified a total of 19,226 InDel loci based on ddRAD-Seq. The distribution of these loci across chromosomes confirmed the suitability of this approach for the development of genome-wide molecular markers. This genome reduction method was also used to develop markers successfully in sesame (Kizil et al. 2020) and lettuce (Seki 2022). However, this study is the first to report the suitability of the ddRAD-Seq approach in the development of InDel markers in sorghum.
Table 1 illustrated the identification of 14 InDel types. InDels that were 10 bp or longer made up 0.4% of all the identified InDels. Generally, as the size of an InDel increased, InDel numbers decreased in the sorghum genome. Furthermore, single-nucleotide InDels were predominant (Table 2), which is consistent with the previous findings in sesame (Kizil et al. 2020), ginkgo (Wang et al. 2023), and soybean (Song et al. 2015). On the other hand, maize (Batley et al. 2003) exhibited a higher prevalence of bi-nucleotide InDels. Our analysis revealed that chromosome 1 had the highest number of InDels with a minimum length of 2 bp. This finding aligns with prior research on sorghum, which indicated that chromosome 1 exhibited the highest abundance of InDels (Li et al. 2016) and SSRs (Shehzad et al. 2020). These findings may also be attributed to the increased likelihood of survival among plants with shorter InDel lengths. Both insertion and deletion events may have negative impacts on the genome, resulting in the loss of some genes and ultimately causing harm to plants. When the length of InDel is shorter, the resulting harm to the plant may be less severe, hence increasing the likelihood of its survival. Thus, the probability of preserving this variation increases (Liu et al. 2019a). Moreover, no deletions and insertions of 10 base pairs or longer were observed on chromosomes 1 and 3, respectively. This could be a drawback of ddRAD-Seq, resulting in large gaps in genome coverage following the sequencing of a prepared genomic library (Shirasawa et al. 2016).
The present study revealed that a majority of the InDel events occurred inside non-coding regions (Table S4), similarly to the findings of Zheng et al. (2011), who developed InDel markers from sweet and grain. The proportions of InDel primers developed from regions of mRNA, introns, exons, and CDS were 12.7%, 6.4%, 4.2%, and 2.1%, respectively (Table S4). InDels, which occur inside functionally significant areas of genes, particularly coding sequence (CDS) regions, have the potential to impact gene function by causing frameshifts and altering the structure of proteins (Zhang et al. 2016). The results of the study indicated that there was a limited presence of InDels inside the CDS (14 bp deletion, position; 47,562,364) area. This result may be attributed to the relatively small proportion of the CDS region represented in the analysis as well as its higher degree of conservation compared to other genomic regions (Liu et al. 2019b). Moreover, InDel within CDS regions has a propensity to have a substantial influence on the structure and function of proteins (Liu et al. 2019b) and can be considered a valuable resource for developing phylogenetic and/or functional markers (Ramakrishna et al. 2018). For example, researchers previously published the identification of the InDel-1891 marker, which has the capability to distinguish restorer lines that possess the Rf1 gene in cotton (Wu et al. 2017).
InDels are the second most prominent kind of structural variation after SNPs uncovered in the plant genome (Yang et al. 2014). These markers have robustness and efficiency comparable to SSRs (Shi et al. 2023) and may be chosen based on their expected fragment size and then confirmed in genetic populations or germplasm via the use of a simple and cost-effective agarose gel electrophoresis technique. Previously, InDel markers have been used for the purpose of analyzing genetic diversity in many crops, including sesame (Kizil et al. 2020), chickpea (Jain et al. 2019), sweet potato (Zhao et al. 2022) and rice (Sahu et al. 2017). However, there are a limited number of studies in which InDel markers have been evaluated for genetic diversity in sorghum (Zheng et al. 2011; Boatwright et al. 2022). In this study, we developed lots of markers that can easily show the polymorphism in agarose gels, and 14 InDel markers were selected to identify genetic variation in 16 sorghum accessions with diverse origins. The polymorphic information content (PIC), which offers an estimation of the information content associated with a marker, is one of the most common measures of the polymorphism of markers (Shete et al. 2000). The analysis of genetic diversity found the highest PIC value was 0.380. The average PIC value of 0.282 for a set of 14 markers is higher than the value reported for the SNP markers used by Yahaya et al. (2023) and Enyew et al. (2022) in sorghum. The results of this research indicated that the InDel markers had a high ability to reveal genetic diversity in the collection. The observed mean values for He (0.07) and I (0.01) were indicative of low genetic variation within the sorghum accessions. In general, previous study on sorghum have shown relatively low genetic variation among landrace accessions (Motlhaodi et al. 2017). The observed result may be attributed to a mix of its inbreeding nature and the rigorous selection criteria used by farmers. In this study, the principal coordinate analysis (PCoA), which explained 26.50% of the total variation in its first two principal axes, provided three class in relation to geographical origin. Accessions were mostly based on the geographical origins in the first and third groups, with some exceptions. This condition may arise from the hybridization of different varieties across continents, facilitating the exchange of cross-gene flows. Moreover, these exceptions indicate that there is a high probability of sorghum landrace genotypes being shared throughout different areas by farmers, perhaps via various means of exchange. This observation is consistent with the idea of seed mixing, exchange, and trade among small-scale farmers, which underscores the dynamic character of genetic connections and seed mobility within agricultural communities.
Conclusion
Next generation sequencing of sorghum accessions with a kind of genome reduction method of ddRAD-seq made possible to develop a large number of InDel markers in sorghum. The findings of this study suggested that the use of this technology is an efficient for the development of genome-wide markers. Among the 80 InDel sites with a minimum length of 10 base pairs, 47 markers that could be resolved using agarose gel electrophoresis were effectively amplified. Due to its ease of detection and cost-effectiveness via the use of readily accessible PCR equipment, these markers may be significant value in genetic analysis and breeding programs pertaining to sorghum.
Data availability
The datasets generated during and/or analyzed during the current study are available in the NCBI repository: https://www.ncbi.nlm.nih.gov/, PRJNA1045240.
References
Ahnert D, Lee M, Austin DF et al (1996) Genetic diversity among elite sorghum inbred lines assessed with DNA markers and pedigree information. Crop Sci 36:1385–1392
Almodares A, Hadi MR (2009) Production of bioethanol from sweet sorghum: a review. Afr J Agric Res 4:772–780
Batley J, Barker G, O’Sullivan H et al (2003) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 132:84–91. https://doi.org/10.1104/pp.102.019422
Boatwright JL, Sapkota S, Jin H, Schnable JC, Brenton Z, Boyles R, Kresovich S (2022) Sorghum association panel whole-genome sequencing establishes cornerstone resource for dissecting genomic diversity. Plant J 111:888–904. https://doi.org/10.1111/tpj.15853
Britten RJ, Rowen L, Williams J, Cameron RA (2003) Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci USA 100:661–4665. https://doi.org/10.1073/pnas.033096410
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:884–890. https://doi.org/10.1093/bioinformatics/bty560
Choe ME, Kim JY, Nabi RBS, Sand-Ik H, Cho KS (2023) Development of InDels markers for the identification of cytoplasmic male sterility in Sorghum by complete chloroplast genome sequences analysis. Front Plant Sci 14:1188149. https://doi.org/10.3389/fpls.2023.1188149
Cui YX, Xu GW, Magill CW, Schertz KF, Hart GE (1995) RFLP-based assay of Sorghum bicolor (L.) Moench genetic diversity. Theor Appl Genet 90:787–796. https://doi.org/10.1007/BF00222013
Doyle JJ, Doyle JL (1990) A rapid total DNA preparation procedure for fresh plant tissue. Focus 12:13–15
Doggett H (1988) Sorghum. Wiley, New York
Enyew M, Feyissa T, Carlsson AS, Tesfaye K, Hammenhag C, Geleta M (2022) Genetic diversity and population structure of Sorghum [Sorghum Bicolor (L.) Moench] accessions as revealed by single nucleotide polymorphism markers. Front Plant Sci 12:799482. https://doi.org/10.3389/fpls.2021.799482
Freese NH, Norris DC, Loraine AE (2016) Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32:2089–2095. https://doi.org/10.1093/bioinformatics/btw069
Gao Q, Yue G, Li W, Wang J, Xu J, Yin Y (2012) Recent progress using high-throughput sequencing technologies in plant molecular breeding. J Integr Plant Biol 54(4):215–227. https://doi.org/10.1111/j.1744-7909.2012.01115.x
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv 1207:3907. https://doi.org/10.48550/arXiv.1207.3907
Gerrano AS, Labuschagne MT, Biljon A, Shargie NG (2014) Genetic diversity assessment in sorghum accessions using qualitative morphological and amplified fragment length polymorphism markers. Sci Agricola 71:345–355. https://doi.org/10.1590/0103-9016-2013-0251
Girardot C, Scholtalbers J, Sauer S, Su S, Furlong EEM (2016) Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinform 17:419. https://doi.org/10.1186/s12859-016-1284-2
Jain A, Roorkiwal M, Kale S, Garg V, Yadala R, Varshney RK (2019) InDel markers: an extended marker resource for molecular breeding in chickpea. PLoS ONE 14:e0213999. https://doi.org/10.1371/journal.pone.0213999
Jones N, Ougham H, Thomas H, Pašakinskienė I (2009) Markers and mapping revisited: finding your gene. New Phytol 183:935–966. https://doi.org/10.1111/j.1469-8137.2009.02933.x
Kedir A, Kebede M, Enyew M et al (2024) Genetic structure and gene flow in Ethiopian sorghum [Sorghum bicolor (L.) Moench] landraces. Genet Resour Crop Evol 71:1241–1256. https://doi.org/10.1007/s10722-023-01687-5
Kimber C (2000) Origins of domesticated sorghum and its early diffusion to India and China. In: Smith CW, Frederickson RA (eds) Sorghum: origin, history, technology, and production. Wiley, New York, pp 3–98
Kizil S, Basak M, Guden B, Tosun HS, Uzun B, Yol E (2020) Genome-wide discovery of indel markers in sesame (Sesamum indicum L.) using ddRADseq. Plants 9:1–1262. https://doi.org/10.3390/plants9101262
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Li J, Wang L, Zhan Q, Liu Y, Yang X (2016) Transcriptome characterization and functional marker development in Sorghum Sudanense. PLoS ONE 11(5):e0154947. https://doi.org/10.1371/journal.pone.0154947
Liu J, Li J, Qu J, Yan S (2015) Development of genome-wide insertion and deletion polymorphism markers from next-generation sequencing data in rice. Rice 8:27. https://doi.org/10.1186/s12284-015-0063-4
Liu N, Wang L, Wu Y et al (2019a) Identification of genome-wide insertion and deletions for sugar beet (Beta vulgaris) using next-generation re-sequencing. Intl J Agric Biol 22:102–106
Liu SR, An YL, Tong W et al (2019b) Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis) and development of InDel markers for genetic research. BMC Genom 20:935. https://doi.org/10.1186/s12864-019-6347-0
Mace ES, Cruickshank AW, Tao Y et al (2020) A global resource for exploring and exploiting genetic variation in sorghum crop wild relatives. Crop Sci 61:150–162. https://doi.org/10.1002/csc2.20332
Mamo W, Enyew M, Mekonnen T et al (2023) Genetic diversity and population structure of sorghum [Sorghum bicolor (L.) Moench] genotypes in Ethiopia as revealed by microsatellite markers. Heliyon 9:12830. https://doi.org/10.1016/j.heliyon.2023.e12830
Mathur S, Umakanth AV, Tonapi VA et al (2017) Sweet sorghum as biofuel feedstock: recent advances and available resources. Biotechnol Biofuels 10:146. https://doi.org/10.1186/s13068-017-0834-9
Medraoui L, Rabeh K, Ater M (2024) Genetic diversity analysis of sorghum (Sorghum bicolor L. Moench) landraces from northwestern Morocco using ISSR and AFLP markers. Genet Resour Crop Evol 71:835–850. https://doi.org/10.1007/s10722-023-01665-x
Motlhaodi T, Geleta M, Chite S et al (2017) Genetic diversity in Sorghum [Sorghum bicolor (L.) Moench] germplasm from Southern Africa as revealed by microsatellite markers and agro-morphological traits. Genet Resour Crop Evol 64:599–610. https://doi.org/10.1007/s10722-016-0388-x
Park SDE (2002) Trypanotolerace in West African cattle and the population genetic effects of selection. Dissertation, Trinity College
Paterson AH (2008) Genomics of Sorghum. Int J Plant Genom 5:1–6. https://doi.org/10.1155/2008/362451
Peterson BK, Weber JN, Kay EH et al (2012) Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135. https://doi.org/10.1371/journal.pone.0037135
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Ramakrishna G, Kaur P, Nigam D et al (2018) Genome-wide identification and characterization of InDels and SNPs in Glycine max and Glycine soja for contrasting seed permeability traits. BMC Plant Biol 18:141. https://doi.org/10.1186/s12870-018-1341-2
Raza A, Ashraf F, Mehmood SS (2019) Evaluation of genetic diversity among exotic Sorghum (Sorghum bicolor L. Moench) genotypes through molecular based analysis (RAPD-PCR). Gesunde Pflanzen 71:187–196. https://doi.org/10.1007/s10343-019-00464-8
Ren Y, Sun X, Nie J et al (2023) Mapping QTL conferring flag leaf senescence in durum wheat cultivars. Mol Breed 43:66. https://doi.org/10.1007/s11032-023-01410-3
Ruiz-Chutan JA, Salava J, Janovska D et al (2019) Assessment of genetic diversity in sorghum bicolor using RAPD markers. Genetika 51:789–803
Sahu PK, Mondal S, Sharma D et al (2017) InDel marker based genetic differentiation and genetic diversity in traditional rice (Oryza sativa L.) landraces of Chhattisgarh, India. PLoS ONE 12:e0188864. https://doi.org/10.1371/journal.pone.0188864
Seki K (2022) Detection of candidate gene LsACOS5 and development of InDel marker for male sterility by ddRAD-seq and resequencing analysis in lettuce. Sci Rep 12:7370. https://doi.org/10.1038/s41598-022-11244-2
Shehzad T, Okuno K (2020) Genetic analysis of QTLs controlling allelopathic characteristics in sorghum. PLoS ONE 15(7):e0235896. https://doi.org/10.1371/journal.pone.0235896
Shete S, Tiwari H, Elston RC (2000) On estimating the heterozygosity and polymorphism information content value. Theor Popul Biol 57:265–271. https://doi.org/10.1006/tpbi.2000.1452
Shi Y, Zhu H, Zhang J et al (2023) Development and validation of molecular markers for double flower of Prunus mume. Sci Hortic 310:111761. https://doi.org/10.1016/j.scienta.2022.111761
Shirasawa K, Hirakawa H, Isobe S (2016) Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res 23(2):145–153. https://doi.org/10.1093/dnares/dsw004
Song X, Wei H, Cheng W, Yang S, Zhao Y, Li X, Feng X (2015) Development of INDEL markers for genetic mapping based on whole genome resequencing in soybean. G3 Genes|Genomes|Genetics 5(12):2793–2799. https://doi.org/10.1534/g3.115.022780
Techale B, Dong H, Mihrete G et al (2022) QTL analysis in multiple sorghum mapping populations facilitates dissection of the genetic control of agronomic and yield-related traits in Sorghum [Sorghum bicolor (Moench)]. Euphytica 218:24. https://doi.org/10.1007/s10681-022-02968-3
Untergasser A, Nijveen H, Rao X et al (2007) Primer3Plus, an enhanced web interface to primer3. Nucleic Acids Res 35:W71–W74. https://doi.org/10.1093/nar/gkm306
Wang D, Zhou Q, Le L, Fu F, Wang G, Cao F, Yang X (2023) Molecular characterization and genetic diversity of Ginkgo (Ginkgo biloba L.) based on insertions and deletions (InDel) markers. Plants 12(13):2567. https://doi.org/10.3390/plants12132567
Wang L, Li J, Zhan Q, Shen Y (2022) Development and application of InDel markers based on sudangrass RAD-seq data. ScienceAsia 48:491–495
Wu J, Zhang M, Zhang X et al (2017) Development of InDel markers for the restorer gene Rf1 and assessment of their utility for marker-assisted selection in cotton. Euphytica 213:251. https://doi.org/10.1007/s10681-017-2043-9
Yahaya MA, Shimelis H, Nebie B et al (2023) Genetic diversity and population structure of African Sorghum (Sorghum bicolor L. Moench) accessions assessed through single nucleotide polymorphisms markers. Genes 14:1480. https://doi.org/10.3390/genes14071480
Yang J, Wang Y, Shen H et al (2014) In silico identification and experimental validation of insertion-deletion polymorphisms in tomato genome. DNA Res 21:429–438. https://doi.org/10.1093/dnares/dsu008
Zhang JZ, Liu SR, Hu CG (2016) Identifying the genome-wide genetic variation between precocious trifoliate orange and its wild type and developing new markers for genetics research. DNA Res 23:403–414. https://doi.org/10.1093/dnares/dsw017
Zhao L, Qi Z, Xiao S, Tang F, Liu Y, Deng Y, Dai X et al (2022) Genetic diversity assessment of sweetpotato germplasm in china using InDel markers. Agronomy 12:3074. https://doi.org/10.3390/agronomy12123074
Zheng LY, Guo XS, He B et al (2011) Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol 12:114. https://doi.org/10.1186/gb-2011-12-11-r114
Acknowledgements
The authors acknowledge the Scientific and Technological Research Council of Türkiye (TUBITAK) for funding this research under the grant number of 118O169.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). This study was supported by the Scientific and Technological Research Council of Türkiye (TUBITAK) with a grant number of 118O169.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Birgul Guden, Ousseini Kiemde, Engin Yol and Bulent Uzun. The first draft of the manuscript was written by Birgul Guden and Ousseini Kiemde. All authors commented on previous versions of the manuscript. Engin Yol and Bulent Uzun read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guden, B., Kiemde, O., Yol, E. et al. Development and utilization of genome-wide InDel markers in Sorghum [Sorghum bicolor (L.) Moench]. Euphytica 220, 75 (2024). https://doi.org/10.1007/s10681-024-03335-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-024-03335-0