Abstract
Trisomic cauliflower plants (Brassica oleracea L. var. botrytis) display abnormal curd phenotypes that seriously decrease commercial value of the crop. Despite extensive breeding efforts, selection of genotypes producing euploid gametes remains unsuccessful due to unknown genetic and environmental factors. To reveal an eventual role of an-euploid gametes, we analyzed chromosome pairing, chiasma formation and chromosome segregation in pollen mother cells of selected cauliflower genotypes. To this end we compared three genotypes exhibiting Low with < 5%, Moderate with 5–10% and High with > 10% aberrant offspring, respectively. Although chromosome pairing at pachytene was regular, cells at diakinesis and metaphase I showed variable numbers of univalents, suggesting partial desynapsis. Cells at anaphase I–telophase II exhibit various degrees of unbalanced chromosome numbers, that may explain the aneuploid offspring. Immunofluorescence probed with an MLH1 antibody demonstrated fluorescent foci in all genotypes, but their lower numbers do not correspond to the number of putative chiasmata. Interchromosomal connections between chromosomes and bivalents are common at diakinesis and metaphase I, and they contain centromeric and 45S rDNA tandem repeats, but such chromatin connections seem not to affect proper disjoin of the half bivalents at anaphase I. Moreover, male meiosis in the Arabidopsis APETALA1/CAULIFLOWER double mutant with the typical cauliflower phenotype does show interchromosomal connections, but there are no indications for partial desynapsis. The causality of the curd development on the desynapsis in cauliflower is still a matter of debate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyploid and aneuploids plants are known to produce significant numbers of trisomics and other numerical variants in their self-fertilized or backcrossed offspring (Sybenga 1975). The most common explanation is that plants with unequal chromosome numbers display unbalanced chromosome segregation during meiotic divisions and so lead to aneuploid gametes (Sybenga 1992). In addition, extra copies of chromosomes in the germ line may also result from incidental mitotic or meiotic non-disjunctions, but these events are rare. An exception is cauliflower (Brassica oleracea L. var. botrytis) in which trisomic descendants can occur in the offspring of diploid progenitors. Such aneuploids with characteristic small and irregularly shaped curds cause considerable economic loss. Despite extensive breeding efforts, selection of genotypes producing euploid gametes is unsuccessful as both genetic, epigenetic, and environmental factors affect regular meiosis with balanced gametes (Chable et al. 2008; 2009).
Unbalanced chromosome segregation in meiosis can be the result of incomplete or no pairing between homologous chromosomes, known as asynapsis and can be caused by genes involved in the upstream part of meiotic recombination machinery, such as ASY1 and SPO11 (e.g., Ross et al. 1997; Wei and Zhang 2010). Such genes are known to disrupt early steps in the pairing initiation and crossover processes and normally lead to complete loss of control of meiosis and gamete degeneration. The second group are the desynapsis mutants, which display regular chromosome pairing or synapsis at early meiosis with an apparently normal tripartite synaptonemal complex, but fail to complete crossover formation, resulting in less or no chiasmata (Ross et al. 1997). Other examples of meiotic aberrations that can lead to the unbalanced segregation of chromosomes, involve merotelic kinetochore attachment mutants in which kinetochores attach to microtubules originating from both spindle poles (Shi and King 2005). Another aberration of chromosome non-disjunction one may consider is the phenomenon in which meiotic or mitotic chromosomes or chromatids do not segregate properly during cell division and hence may result in aneuploidy. When mitotic non-disjunction of individual chromosomes occurs, it may create mosaicism of euploid and aneuploid germlines, and may cause, an increased rate of aneuploidy in eggs or sperm (Robinson and McFadden 2002). More meiotic disturbances that may give rise to aneuploidy or changes in ploidy are (aspecific) stickiness, premature loss of sister-chromatid cohesion, failure of cytokinesis, microtubule errors, and first and second division restitution (FDR, SDR), and were described comprehensively in reviews of Arabidopsis thaliana (Castellano and Sablowski 2008; Peirson et al. 1997; Yang et al. 1999; Zamariola et al. 2014); Brassica napus (Souza and Pagliarini 1996), wheat (Huskins and Hearne 1933), maize (Beadle 1937, 1933a,b; Caetano-Pereira et al. 1995), tomato (Soost 1951) and others. Unbalanced gametes may give rise to aneuploid offspring, as was demonstrated for various crops and model species including Arabidopsis (Grelon et al. 2003), wheat (Griffiths et al. 2006) and maize (Carlson et al. 2007).
In this study, we present an analysis of meiosis in pollen mother cells in cauliflower with special attention on disturbances that give rise to aneuploid spores. We compare the course of meiotic states in three inbred lines differing in the percentage of aneuploid offspring. Our observations suggest that partial desynapsis is the most likely mechanism leading to less chiasmata, and hence to univalents at diakinesis/metaphase I, and so to more unbalanced chromosome segregation at later stages, and as we assume to higher frequency of aneuploid gametes. We also noticed abundant interchromosomal connections and asked the question whether this relates to decreased number of chiasmata and more univalents. In addition, we speculated that allelic variants of genes involved in curd formation itself can, as a side effect, induce erratic meiosis. To this end we investigated meiotic stages in the pollen mother cells of the APETALA1/CAULIFLOWER double mutant of Arabidopsis thaliana (Bowman et al. 1993; Smyth 1995) with the typical cauliflower like curd morphology.
Material and methods
We selected three genotypes of cauliflower (B. oleracea L. var. botrytis) that differ in frequency of aneuploid individuals in their offspring. Aneuploid individuals among offspring were identified by their characteristic aberrant phenotypes and leaf samples of these plants were used for flowcytometric measurements of its genomic DNA. Part of the material was also karyotyped based on repeat and BAC painting, confirming trisomy or telo-trisomy (Ji 2014). The so-called Low genotype is an inbred line with 68 days of growing and produces less than 5% aneuploids; the Moderate genotype is an inbred line with 77 days of growing and has 5–10% aneuploids; the High genotype is an inbred line with 80 days of cultivation and more than 10% aneuploids. All cauliflower lines were bred for optimal performance under temperate climate conditions. The APETALA1/CAULIFLOWER double mutant of Arabidopsis thaliana with a cauliflower-like curd was obtained from Dr. Kerstin Kaufmann of Bioscience, Plant Research International, WUR, Wageningen (Bowman et al. 1993).
Repetitive DNA sequence for specific chromosome painting
The plasmid pTa71 containing the 45S rDNA repeat (Gerlach and Bedbrook 1979) was isolated with the High plasmid purification kit of Roche (REF: 11754785001). The Brassica rapa centromere specific repetitive sequences CentBr1 and CentBr2 were previously shown to paint centromere regions of B. oleracea L. var. botrytis (Lim et al. 2005, 2007; Xiong and Pires 2011). These centromere-specific repetitive sequences were PCR amplified using the following primer sets:
CentBr1.
Forward primer: 5′-GAATAGCACAGCTTCATCGTCGTTCC-3′.
Reverse primer: 5′-CTGGGAAACTGTAATCACCTGATCTGAAA-3′.
CentBr2.
Forward primer: 5′-GGGAATATGACACCTTCTTTGTCATTCT-3′.
Reverse primer: 5′-CAGGAAAACTGGGATCACCTGATTTAAAT-3′.
Slide preparation
Young flower buds of cauliflower and APETALA1/CAULIFLOWER double mutant of Arabidopsis thaliana were fixed in freshly prepared ethanol: acetic acid (3:1) at 20 °C and after one day transferred to ethanol 70% for longer storage at 4 °C. Anthers were dissected from the flower buds, washed in purified water two times and in 10 mM Na-citrate buffer (pH 4.5), respectively, and then mildly digested in a pectolytic enzyme mix, containing cellulase RS (Yakult Pharmaceutical IND.CO, LTD, Tokyo, Japan, Yakult 203033), pectolyase Y23 (from Aspergillus japonicus, Sigma Aldrich, St. Louis, MO, USA, P-3026) and cytohelicase (from Helix pomatia, Sigma Aldrich, St. Louis, MO, USA, C8274) in the Na-citrate buffer at a 0.3% final concentration for each. The flower buds were macerated for 3 h at 37 °C in the enzyme solution. The maceration was followed by two washing steps in Milli-Q water. For each preparation we carefully transferred a single anther to a clean slide and dissected the tissues with fine needles to make a cell suspension in 15 µL Milli-Q water. After adding 25 µL 50% acetic acid the cells on the slide were heated for 4 min on a 50 ˚C hot plate. Then, 50 µL ethanol: acetic acid (3:1) fixative was dropped around and on top of the cell suspension, after which the slides were left to dry for few minutes. We finally analyzed the slides under a phase contrast microscope for sufficient spreading and quality (Kantama et al. 2017).
Fluorescence in situ hybridization
We labelled the 45S rDNA probe directly with Di-ethylaminocoumarin-5-dUTP (DEAC, Perkin Elmer Life Sciences, Boston, MA, REF: NEL-455001EA), the Centbr1 probe was labelled with the Biotin-Nick-translation for Cy5 detection (Roche Applied Science, REF: 11745816910) and for the Centrbr2 probe we labelled indirectly with the Dig-Nick-translation for FITC detection (Roche Applied Science, REF: 11745824910) as descripted previously (Kato et al. 2004). Probes of three repetitive sequences were hybridized to various slides of the same genotype. Cell spreads were pre-treated with 1% formaldehyde for extra fixation (10 min at 20 °C), followed by an RNAase treatment (100 μg/mL DNase-free ribonuclease A stock solution, AppliChem, St. Louis, MO, USA, diluted as 1:100 in 2 × SSC (Saline-sodium citrate buffer, pH 7) at 37 °C for 1 h, and then washed again in 2 × SSC for 3 × 5 min. Then slides were fixed in 1% formaldehyde at room temperature for 10 min. After fixation, slides were washed with 2 × SSC and dehydrated in an ethanol series (70%, 90%, and 100% three minutes each).
Ten µL of each DNA probes was added to 10 µL of the hybridization mixture containing 50% formamide, 20% dextran sulphate, followed by denaturation in boiling water for 10 min and put on ice before being added onto the slides. To each slide we applied 20 µL probe mixture, which then was transferred to an 80 °C hot plate for 3 min denaturation. This was followed by overnight hybridization in a humid chamber at 37 °C. After hybridization, slides were washed at 42 °C in 50% formamide/2 × SSC for 3 × 5 min, followed by 3 × 5 min washes in 2 × SSC. For the detection step we amplified the probe with 500 μg/mL biotinylated-anti-streptavidin (Cy-5) (Vector laboratories, BA-0500, stock solution 1:200 diluted in TNB buffer (0.1 M Tris–HCl pH 7.5, 0.15 M NaCl, 0.5% blocking reagent, Roche Applied Science, REF: 11096176001) or 200 μg/mL Anti-digoxigenin-fluorescein (FITC) (Roche, 11207741910) a stock solution diluted as 1:200 in TNB buffer) and signal amplification (Cy-5) (200 μg/mL streptavidin, Alexa Fluor 647 (Invitrogen, S21374) a stock solution diluted as 1:800 in TNB buffer or 200 μg/mL Rabbit-anti-sheep-fluorescein (FITC, Jackson, Bio-connect; 313-096-003) a stock solution diluted 1:800 in TNB buffer. After the detection steps the slides were dehydrated through an ethanol series (70%, 90%, and 100%, three minutes each). The air-dried slides were counterstained with 12 μL of a 300 ng/mL DAPI (4′, 6-diamidino-2-phenylindole, Sigma Aldrich, St. Louis, MO, USA, D-1388) in Vectashield® antifade mounting medium (Vector Laboratories) and covered with a 24 × 50-glass cover slip. The cells were examined with Zeiss Axioplan 2 imaging photomicroscope, equipped with epi-fluorescence illumination and filter sets for DAPI, FITC, Cy5 and DEAC. The images were processed with Genus Image Analysis Workstation software (Applied Imaging) and selected images were captured by a Photometrics Sensys monochrome 1305 × 1024-pixel CCD camera. Different fluorescent signals were captured and combined in Adobe Photoshop by using overlapping layers, gradient color mapping and screen blend mode (Kantama et al. 2017).
Immunodetection of MLH1
For the detection of the mismatch repair protein MLH1, class I crossover protein, we performed immunofluorescence microscopy with primary antibodies for this protein on air-dried acetic spread pollen mother cells at diakinesis. The MLH1 polyclonal antibody of Arabidopsis thaliana was obtained from Dr Liudmila Chelysheva (INRA, Versailles, France) following Chelysheva et al. (2010) with few adaptations. To this end we heated 10 mM Na-citrate buffer (pH 6.0) in a microwave oven at 450 W for 45 s until boiling, submerged the slides in the boiling solution and cooked this in the microwave at 450 W for 45 s. The slides were then transferred to PBS-T buffer (1× Phosphate buffered saline, 0.1% v/v Triton, pH 7.4) and incubated for 5 min, followed by incubation of the slides with the MLH1 polyclonal antibody in PBS-T-BSA (1× PBS, 1%w/v BSA, 0.1% v/v Triton) in a dilution of 1:200 at 4 °C overnight or over the weekend in a humid chamber. After incubation slides were washed in PBS-T for 3 × 15 min. The secondary antibody Fluorescein (FITC)-conjugated AffiniPure F (ab’)2 Fragment Goat Anti-Rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories, Inc.) was diluted in PBS-T-BSA (1:100) and used on the slides for 1 h at 37 °C. Slides were then washed for 3 × 10 min in PBS-T, and then air-dried. Finally, the slides were counterstained in 300 ng/mL DAPI in Vectashield and covered with a 24 × 50-glass cover slide. Microscopy and image processing followed the description above. We pseudo-colored the DAPI image in red for better contrast with the green MLH1 foci.
Statistical analyses of the numerical data
For the diakinesis/metaphase I cells we scored for number of rod and ring bivalents, univalent pairs, chiasma bonds and chromosomes involved in interconnections. Basic statistical analyses were performed in Microsoft Excel®, GenStat v. 16.2 (VSN International Ltd, http://www.vsni.co.uk/software/genstat) and JASP v. 0.14.1 (https://jasp-stats.org/getting-started-with-jasp/).
Results
Meiotic aberrations in the three genotypes
The basis of our analysis is the comparison of morphological and quantitative meiotic characteristics between three cauliflower genotypes that differ in the frequencies of offspring with aberrant phenotype. The so-called Low phenotype produces less than 5% aberrant offspring, the Moderate phenotype between 5 and 10% and the High phenotype more than 10%. For each genotype we studied around 50 DAPI stained pollen mother cells (PMCs), at stages from pachytene to anaphase II, of which the majority were at diakinesis (at least 50 PMCs per genotype were studied).
Figure 1a, b show representative examples of meiotic prophase cells at pachytene. Chromosomes at this stage in all genotypes are fully paired and display the typical pattern of brightly fluorescing pericentromeres, that are often tightly clumped forming a dense synizetic knot (Moens 1964), which we consider artefact of the acetic fixation (Armstrong et al. 2001). The distal parts of the chromosome arms are the less condensed euchromatin with various smaller chromomeres. Although some nuclei display short unpaired stretches, we consider pachytene complements fully synaptic. Incidentally we detect associations between non-homologous centromeres or pericentromeres in the three genotypes (Fig. 1a).
Photomicrographs of DAPI stained pollen mother cells of cauliflower, B. oleracea L. var. botrytis, 2n = 2x = 18, at different meiotic stages. Bar equals 10 μm in the figures. a Cell at pachytene of the High genotype. The homologous chromosomes are clearly almost fully paired. The cells show the typical wild type pachytene morphology with brightly fluorescing pericentromere regions, which cluster at some places (arrows). The weaker fluorescing euchromatin has the characteristic pattern of smaller chromomeres. b Cell of the High type at late pachytene/early diplotene, where only Nucleolar Organizer Region and few other chromosome regions have disjoined (arrow heads)
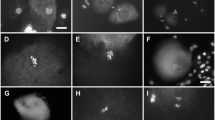
At diakinesis, chromosomes are highly condensed, and pairs kept together by clearly distinguishable chiasmate bonds (Fig. 2a-e). For the interpretation of bivalent configurations, we assume that arms have one chiasma, so the rare case of two adjacent chiasmata in one arm will be interpreted as a single event. Without at least one chiasma, homologues are not attached and so remain separated as a univalent pair (Fig. 2c). With only one chiasma between homologues, bivalents attain the shape of a cross or that of a rod, depending on the position of the chiasma (Fig. 2a). With two chiasmata, a bivalent exhibit distinguishable ring shapes depending on its position, i.e., close to centromere or distal end (Fig. 2b). Most cells contained ring and rod bivalents and univalent pairs (Fig. 2 d, e). In the Low genotype, univalents are rare and occur in only 10.5% of the cells. In the Moderate and High genotypes univalents are much more frequent (39% and 42.3% respectively). DAPI fluorescing interchromosomal connections between the homologous and non-homologous chromosomes are seen in all genotypes. Even in some cells of the Moderate and High genotypes, almost all chromosomes are connected by these threads (Fig. 2f).
DAPI stained pollen mother cells at diakinesis—anaphase II. The figures a and b are from the “Low genotype material; the figures (c), (d) and (i) are from the Moderate genotype; the figures (e), (f), (g), (h) are from the High phenotype. (a) Cell at diakinesis—metaphase I with nine bivalents. Ring-bivalents have at least one chiasma on both chromosome arms. Open rings have distal chiasmata on both arms; compact rings have chiasmata closer to the centromeres. Rod-bivalents are considered to have (mostly one) chiasma on either of the chromosome arms (example of a rod bivalent is shown). b, c Cell with clear examples of interchromosomal connections (arrows). “p” refers to chiasma that is more proximal (closer to the centromere), and “d” more distal. d Cell with two univalents, of which apparently one is associated with a nearby bivalent. e In this cell two bivalents are tightly connected, giving the impression of a quadrivalent. f Metaphase I cell with two univalent pairs (I-pairs) shown with thin arrows. g Anaphase I cell with 10 visible univalents of which one may be a univalent chromatid. h Anaphase II showing two lagging chromosomes with interconnection. i Anaphase II cell with one lagging chromosome. Two of the anaphase II complements are on top of each other. The segregation of 8 + 9 + 1 + 18 chromatids is clearly unbalanced
At metaphase I bivalents transgress to the equatorial plate with centromeres facing the poles. In a few cases, we observed multivalent-like configurations due to interchromosomal connections as mentioned above (Fig. 2f). At anaphase I, the half bivalents disjoin normally, whereas the univalents segregate un-equal resulting in unbalanced chromosome numbers at later stages. Moreover, anaphase bridges and multiple lagging chromosomes appeared (Fig. 2g) in the Moderate and High genotypes. At anaphase II, we observed anaphase bridges and interchromosomal connections (Fig. 2g, h). In cells at the tetrad stage, we found mostly balanced tetrads (9 + 9 + 9 + 9) in the Low genotype, while in the Moderate and High genotypes, tetrads with unbalanced chromosome numbers like (9 + 9, 9 + 8 + 1) and (10 + 8, 10 + 8) were observed (Fig. 2i).
Number of chiasmate bonds in the three genotypes
Since the Moderate and High genotypes demonstrate relatively high numbers of non-chiasmatic chromosomes, we estimate the total number of chiasmata based on the numbers of counted rod and ring bivalents (with the assumption that they account for 1 and 2 chiasmata, respectively, Table 1, Fig. 3b). Chiasma estimates for the Low, Moderate and High genotypes were 14, 13 and 14 per cell respectively, which is statistically not significant (P = 0.152). Hence, there is no clear relation between the number of chiasmata bonds and the number of aneuploid offspring.
a Pie charts presenting the proportions of ring bivalents, rod bivalents and univalent pairs in the Low, Moderate and High genotypes of cauliflower, B. oleracea L. var. botrytis, 2n = 2x = 18. b Box and whisker plots of the number of chiasmata bonds of the Low, Moderate and High genotypes. c Scatter plot showing number of interchromosomal connections and number of chiasmata bonds for the Low, Moderate and High genotypes. The trendlines for each of the genotypes demonstrates that interchromosomal connections and number of chiasmata are statistically not significant
We therefore compared the mean number of estimated chiasmata in cells with univalent pairs with those that lack univalents. Our data suggests that univalent pairs lead to increase of chiasmata on other chromosome pairs within the cell complement (Table 2).
More univalents in the Moderate and High genotypes
Numbers and frequencies of univalent pairs, rod bivalents and ring bivalents at diakinesis cells for the three genotypes are summarized in Table 1. The percentages of cells showing an expected number of nine bivalents are 89%, 61% and 57% for the Low, Moderate and High genotypes, respectively. In the Low genotype, 7% of the cells have one pair of univalents and 4% of cells have two pairs of univalent. In the Moderate genotype, 27% of the cells have one pair of univalents and 12% of cells have two pairs of univalents. In the High genotype, 31% of the cells have one pair of univalents and 12% of cells have two or more pairs of univalents. To determine whether the genotypes differ significantly in univalent incidence, we performed a one-way ANOVA test on univalent counts showing significant differences between the three genotypes (P = 0.002) (Fig. 3a). In the Fisher's protected least significant difference test (series of pairwise t-tests) we showed that the Low genotype differs significantly from both the Moderate and High genotypes, while the Moderate and the High genotypes do not differ from one another (Table 3).
Immunodetection of chiasma precursor MLH1
We further tested the nature of the chiasmata bonds with an immunofluorescence detection on spread diakinesis cells using antibodies against the chiasma precursor MLH1. MLH1 is known to reside in the later recombination nodules of the synaptonemal complex at pachytene and is involved in the class I crossover pathway. The MLH1 foci in our slides clearly correspond to most of the chiasma sites (Fig. 4) but are lacking in some places that may represent sites of class II crossovers or class I crossover sites that were not detected by the immune fluorescent method. We analyzed at least 15 cells for each genotype showing on average 10.93, 10.43 and 9.52 MLH1 sites per cell in the Low, Moderate and High genotype, respectively. Compared to the number of chiasmata bonds, we calculated that MLH1 stained chiasmata amounted 77.5% for the Low and the Moderate type, and 69% for the High genotype (Table 1).
FISH probed with labelled repetitive sequences on spread pollen mother cells at diakinesis of the High genotype. a and c 2-colour FISH with the DAPI imaging and the blue 45S rDNA foci; b and d 4-colour FISH with the red fluorescence signals of the Brassica specific centromere repeat Centbr1 (pseudo-colored in red) and green fluorescence of the Brassica specific centromere repeat Centbr2. The arrows point at connections between chromosomes that colocalize with the 45S rDNA and centromere repeats
Interchromosomal connections
The presence of bivalent connections is a second remarkable feature in meiosis of these cauliflower genotypes. We counted numbers of connections between bivalents (Table 1). The average numbers of connections were 1.1, 2.0 and 2.1 in the Low, Moderate, and High genotypes, respectively. A one-way ANOVA demonstrated that differences in bivalent interconnections between the three lines are significant (P-value < 0.001), and a subsequent Fisher's protected least significant difference test indicates that the Low genotype differs significantly from both the Moderate and High genotype (P < 0.001). However, the difference between the Moderate and High genotype is not significant (0.5286). Comparison of the three lines for the correlation of numbers of crossovers and numbers of interchromosomal connectives (Fig. 3c) revealed that both variables were not significant (Kendall’s Tau coefficient: Low = 0.982, Moderate = 0.834, High = 0.375).
Repetitive DNA FISH analysis of diakinesis/metaphase I
To investigate if interchromosomal connections contain repetitive sequences, as was previously suggested for Ornithogalum longibracteatum (Pedrosa et al. 2001), we performed a FISH experiment with the 45S rDNA and two Brassica-specific centromere repeats as probes. The results indicate that 45S rDNA as well as the two Brassica repeats painted interchromosomal connections (Fig. 5a, c). Centromere and 45S rDNA specific sequences involved around 70% of bivalent connections, while less than 30% of the connections comprised non-stained chromatin. When 45S rDNA connections were observed, one or both chromosomes involved carry a 45S rDNA locus, whereas when centromere repeats are involved, these connect the centromere regions of two homologues (Fig. 5b, d). We then asked as to whether satellite (NOR) chromosomes are more often involved in interconnections. Two of the nine chromosome pairs have 45S rDNA loci. Connections between chromosomes with 45S rDNA (45S-45S), between a chromosome with and without a 45S rDNA signal (45S-non) and between chromosomes without 45S rDNA signals (non- non) were quantified for a subset of 19 painted interconnections (see Table 4). The theoretical chances of connections between chromosomes (while assuming no preferential connections being formed) are given by the following chances: P(45 s-45 s) = 2/9*1/8 = 1/36, P(45S-non) = 2/9*7/8 + 7/9*2/8 = 14/36 and P(non-non) = 7/9*6/8 = 21/36. A computed χ2 test of 27.8, and pdf = 2 < 0.001, clearly shows a significant overrepresentation of 45S rDNA repeats in interchromosomal connections (Table 5). At least for the 45S rDNA, the interconnections are not random, but preferentially occur between chromosomes that have functionally similar regions.
Immunofluorescence with the MLH1 antibody on diakinesis cells of a Low type, b Moderate and c High genotype. MLH1 foci demonstrate sites of late recombination nodules, the place of crossover events. The number of MLH1 foci is lower than expected possibly due to inaccessible target sites. For better contrast DAPI images have been pseudocolored in red
Meiotic analysis of the APETALA1/CAULIFLOWER Arabidopsis mutant
Since cauliflowers are known for significant numbers of trisomic offspring, we wondered whether genes responsible for curd formation indirectly cause more cells with partial desynapsis. We therefore studied meiotic stages of the APETALA1/CAULIFLOWER double mutant of Arabidopsis that, like Cauliflower, displays the typical curd phenotype (Fig. 6a). Microscopic observations of pollen mother cells at diakinesis in this double mutant showed interchromosomal connections (Fig. 6b, c), albeit far less than in Brassica: in less than 10% of the analyzed cells. Univalents were not observed neither did these plants produce aneuploid offspring. We conclude that APETALA1 (AP1) and CAULIFLOWER (CAL) genes do not cause partial desynapsis during meiosis stages in Arabidopsis.
a The AP1/Cau double mutant of Arabidopsis forms a typical curd phenotype resembling that of cauliflower. b and c show two photomicrographs of meiotic pollen mother cell of the AP1/Cau mutant at diakinesis. Interconnections between bivalents are clearly observed (arrows) whereas univalent pairs are completely lacking
Discussion
Crossover recombination
In this study we aimed to explore whether aneuploidy in cauliflower (Brassica oleracea L. var. botrytis) progeny is correlated with disturbances in chromosome pairing, recombination, and segregation during meiosis. The underlying assumption was that aneuploid gametes are the most likely source of aneuploids in the offspring. We therefore analyzed male meiosis in spread pollen mother cell preparations of three cauliflower genotypes, Low, Moderate and High, that differ in percentages of aneuploid offspring. All three genotypes display regular chromosome pairing at pachytene, while cells at diakinesis – metaphase I can contain one or few univalent pairs, a phenomenon known as partial desynapsis. We observed that in the Low and High genotypes, even 40% of cells contain such univalent pairs. Most of the cells show one univalent pair, but in the Moderate and High genotypes we often saw more univalent pairs. Univalents that move to either pole or remain lagging in the equatorial cell plane may lead to unbalanced chromosome numbers at tetrad stage. Only spores with 9 or 10 chromosomes remain (spores with 8 chromosomes abort), of which the latter chromosome number may give rise to offspring plants with aberrant aneuploid phenotype.
The number of univalent pairs in the male meiocytes is clearly higher in the Moderate and High genotypes, but the difference between these numbers is not reflected by the differences in aberrant offspring between the Moderate and High type cauliflowers. We know that sample size of the analyzed meiotic cells is relatively small as slides with high quality cell spreads at different meiotic stages is time consuming and laborious. In addition, we have not been able to account for eventual effects of developmental, epigenetic, and environmental conditions that may influence the incidence of univalents. We also lack a non-desynaptic cauliflower as a control, so cauliflower plants with normal meiosis and balanced chromosome segregation and euploid gametes. However, based on recent genetic studies, we can predict the number of chiasmata derived from estimated genetic map lengths for a Brassica oleracea with euploid offspring. According to Zhao et al. (2016) the total map length of cauliflower is estimated at 890 cM, which corresponds to an estimated crossover event number of 890/50 = 18 per diploid cell complement, which equals on average two crossover events per chromosome. Yu et al. (2019) found comparable values for broccoli, which has an estimated map length of 799 cM, which equals 16 per cell complement and 1.78 c.o. per chromosome, respectively. These estimates are slightly higher than the partial desynaptic genotypes in our study. The slightly lower chiasma estimates here can partly be explained by incidental double chiasmata which have been interpreted as a single chiasma event. A second point of attention is that, for practical reasons, we cannot include a quantitative study of female meiosis as this kind of analyses is extremely laborious and time consuming. We realize that most of the aneuploid gametes get lost in competition to euploid gametes in its way through pollen tuber to egg cell, a process known as certation (Heribert-Nilsson 1920; Koornneef and Van der Veen 1983). This selection mechanism is known to reduce male aneuploid gamete transmission to values often far below 5%. It is therefore likely, that a significant rate of the aneuploids that we observed in the offspring families originated from aneuploid female gametes.
As basic processes in chromosome pairing, recombination and segregation are the same between male and female meiosis, apart from some recombination frequency differences between male and female meiosis (de Vincente and Tanksley 1991; Drauoud et al. 2007), we nonetheless consider partial desynapsis the primary cause of irregular chromosome segregation in cauliflower. The question now arises as to whether univalent pairs always involve the same chromosomes. Chromosome painting with pooled BACs of Arabidopsis thaliana on mitotic chromosome spreads of this material support the observation that different chromosomes are involved (Ji, 2014).
We further elaborated on the number of crossover events in a series of MLH1 immunolabelling experiments on cell spreads of the Low, Moderate and High genotypes. MLH1, which is involved in the ZMM interference dependent pathway, produce class I crossovers, resides in the late recombination nodules of the synaptonemal complex at pachytene and remains a detectable protein in chiasmata until diakinesis /metaphase I. The immunofluorescent foci revealed that 77.5%, 77.5% and 69% of the chiasmata of the Low, Moderate or High genotypes are MLH1 positive (Table 1). The MLH1 negative chiasmata are supposed to be class II crossover events or are supposed to be chiasmata in which the MLH1 could not be detected. The frequencies of these MLH1 foci correspond well with the estimated frequencies of 80–85% in Arabidopsis (Copenhaver et al., 2002; Chelysheva et al. 2010) and 70% in tomato (Lhuissier et al., 2007).
The relation between the number of univalent pairs and average number of chiasmata bonds and MLH1 foci per cell was surprising. While the Moderate and High genotypes display almost four times more univalents, the average number of chiasmata bonds remained largely the same (Table 1). If we compare the number of chiasma bonds in cells with and without univalents we see a much higher number of chiasma bonds in cells with univalents than in cells without (Table 2), suggesting that chromosome pair(s) that lack the obligate crossover compensate specific processes of crossovers placements in trans, thus keeping the total number of chiasmata in the cell the same, irrespective univalents or not. Apparently, reduction in crossing overs in one or more chromosomes is accompanied by a concomitant increase of crossovers in other chromosomes in the cell, a phenomenon described for partial asynaptic inbred lines of maize and reviewed by Sinha and Mohapatha (1969). The authors argue that a counteracting interchromosomal chiasma compensating mechanism is driven by chiasma interference, which is explained by assuming a definite quantity of material that is essential to chiasma formation in that cell. Carlton et al. (2006) proposed that meiotic progression is delayed on a per-nucleus basis until key events such as recombination have been completed. The apparent action of this checkpoint between early and late pachytene suggests that this transition is a target for cell cycle regulatory machinery. Termolino et al. (2019) described a comparable phenomenon for a paracentric inversion heterozygote in Arabidopsis where crossover suppression in the rearranged chromosome segment is associated with a significant increase of crossovers in the other chromosomes, thereby maintaining the total number of crossovers per cell unaffected. Such a trans regulation mechanism was found restricted to only male meiosis. In chromosome rearrangements of Drosophila, a similar interchromosomal compensation was observed causing a pachytene delay and hence influenced the pachytene checkpoint activity by extending to make more crossovers (Joyce et al. 2011).
Interchromosomal connections
The second striking feature of the partial desynaptic cauliflower genotypes are the interchromosomal connections that were most noticeable in diakinesis and metaphase I cells. We found in each cell at least one of these chromatin threads, while in the Moderate and High genotypes on average two of such connections were observed. Such chromatin threads were different from the non-homologous centromere fusions seen at pachytene, a phenomenon that resembles the previously described non-homologous association in meiotic prophase I chromosomes of Beta vulgaris and are regular features of pachytene chromosomes (de Jong and Stam 1985). Moreover, we did not find MLH1 foci on these threads suggesting that such chromatin threads are not part of distal euchromatin segments, where chiasmata are located. In a series of chromosome painting experiments with FISH using probes of 45S rDNA and centromere repeats, we were able to demonstrate that the Brassica specific pericentromere repeats and the 45S rDNA repeat are involved, and that the NOR chromosomes containing tandem arrays of this repeat display more connections than expected by chance. Furthermore, when centromere sequences were involved, they always connect the pericentromere repetitive regions of different chromosome pairs. An explanation for such connections is still not clear.
The chromosome connections, sometimes referred to as chromosome stickiness in the literature have been described in interspecific hybrids and inbred lines and have been suggested the result of genetic mutations or environmental factors, which leads to unequal distribution of genetic material to the daughter cells (Klášterská and Natarajan 1975, Rayburn and Wetzel 2002). Other reports claim that such interconnections give rise to a higher frequency of non-disjunction, unbalanced segregation at anaphase, increase the frequency of translocations and the rate of gene mutation (Beadle, 1937), abnormal chiasma formation (Higgins et al. 2005) and lead to dicentric anaphase bridges (Basi et al. 2006). Higgins et al. (2011) elucidated that the RecQ helicase AtRECQ4 in Arabidopsis is required for normal levels of fertility, by dissolving recombination intermediates between non-self telomeres during meiosis, and its absence leads to chromatin bridges and chromosome fragmentation in a small number of cells.
Koul and Nagpal (2016) observed inter-telomeric connections in an achiasmatic meiosis in Tradescantia spathacea, in which the connections were prominent at early stages but disappeared at the time of diplotene. It was proposed that alteration in the cellular microenvironment may have altered the telomeric architecture that not only impeded the chiasmata formation but also induced ectopic pairing through inter-telomeric connections. Also, in our cauliflower material it is tempting to assume that interchromosomal connections are the results of ectopic pairing and/or erroneous DNA repair as meiotic double strand breaks in tandem repeat chromosome regions. That also the APETALA1 – CAULIFLOWER double mutant of Arabidopsis demonstrates such interchromosomal connections, albeit at only 10% of the cells, suggests that the cauliflower curd phenotype may leads in some way to cell stress resulting in local ectopic pairing and DNA repair anomalies, but that the disturbed physiology does not lead to chromosome pairing and chiasma problems.
Features and inheritance of desynapsis
Desynaptic mutants belong to the most common synaptic disturbances in higher plants and have been described for many species (reviews in Koduru and Rao, 1981; Kaul and Murthy, 1985; Wani and Bhat, 2017). Most of these mutants were found in natural populations, others were induced by ionizing radiation (Konvička and Gottschalk, 1971; Basi et al. 2006), EMS treatment, were selected in a T-DNA (Peirson et al. 1997) or transposon tagged populations (Cai and Makaroff 2001). In general, desynaptic mutants inherit as a single recessive mutant gene (Beadle 1930; Menzel and Brown 1955), sometimes by multiple genes (Rees, 1961) and were often selected from a group of plant that exhibit male sterility or vary largely in the shape and size of euploid, aneuploid and unreduced pollen grains. The majority of the desynaptic mutants affect the entire chromosomal complement but a few of them affects only specific chromosomes. A striking phenomenon found in desynaptic plants is the occurrence of chromosome stickiness or interchromosomal connections (Rao et al. 1990; Pagliarini et al. 2000; Mehra and Rai 1972; Klášterská et al. 1976; Higgins et al. 2005), although their appearance differs between the species where they were observed. Rao (1975) found that Coix lacryma-jobi demonstrated sterility by desynapsis only during the hot summer months and was supposed the result of genotype-environment interactions. The latter and various other papers suggest that a complex of physiological, developmental and/or epigenetic mechanisms are involved in the loss of meiotic control leading to desynapsis.
Abbreviations
- AP1:
-
APETALA1
- BSA:
-
Bovine Serum Albumin
- CAL :
-
The gene CAULIFLOWER
- Cy5:
-
Cyanine 5 fluorochrome
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DEAC:
-
7-Diethylaminocoumarin-3-carboxylic acid
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- MLH1:
-
mutL homolog1
- PBS-T:
-
Phosphate buffered saline with 0.1% v/v Triton
- PMCs:
-
Pollen mother cells
- SSC:
-
Standard Saline Citrate
- TFL1:
-
TERMINAL FLOWER1
- TNB:
-
Tris–HCl pH7.5, 0.15 M NaCl, 0.5% blocking reagent
References
Armstrong SJ, Franklin FC, Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114(Pt 23):4207–4217
Basi S, Subedi LP, Gopal Bahadur KC, Adhikari NR (2006) Cytogenetic effects of gamma rays on Indica Rice Radha-4. J. Inst. Agric. Anim. Sci. 27:25–36
Beadle GW (1930) Genetical and cytological studies of Mendelian asynapsis in Zea mays. Cornell Univ Agr Expt Sta Mere 129:1–23
Beadle GW (1933a) Further studies of asynaptic maize. Cytologia 4:269–287
Beadle GW (1933b) A gene for sticky chromosomes in Zea mays. Z Indukt Abstamm Vererbungsl 63(1):195–217
Beadle GW (1937) Chromosome aberration and gene mutation in sticky chromosome plants of Zea mays. Cytologia 1:43–56
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Caetano-Pereira CM, Pagliarini MS, Brasil EN, Martins EN (1995) Influence of aluminum in causing chromosome stickiness in maize microsporocytes. Maydica 40:325–330
Cai X, Makaroff CA (2001) The dsy10 Mutation of Arabidopsis results in desynapsis and a general breakdown in meiosis. Sex Plant Reprod 14(1–2):63–67
Carlson SR, Rudgers GW, Zieler H, Mach JM, Luo S, Grunden E, Krol C, Copenhaver GP, Preuss D (2007) Meiotic transmission of an in vitro-assembled autonomous maize minichromosome. PLoS Genet 3(10):1965–1974
Carlton PM, Farruggio AP, Dernburg AF (2006) A link between meiotic prophase progression and crossover control. Plos Genet 2(2):e12. https://doi.org/10.1371/journal.pgen.0020012
Castellano MM, Sablowski R (2008) Phosducin-like protein 3 is required for microtubule-dependent steps of cell division but not for meristem growth in Arabidopsis. Plant Cell 20(4):969–981. https://doi.org/10.1105/tpc.107.057737
Chable V, Rival A, Cadot V, Boulineau F, Salmon A, Bellis H, Manzanares-Dauleux M (2008) “Aberrant” plants in cauliflower: 1. Phenotype and heredity. Euphytica 164:325–337
Chable V, Rival A, Beulé T, Jahier J, Eber F, Cadot V, Boulineau F, Salmon A, Bel-lis H, Manzanares-Dauleux M (2009) ‘“Aberrant”’ plants in cauliflower: 2. Aneuploidy and global DNA methylation. Euphytica 170:275–287
Chelysheva L, Grandont L, Vrielynck N et al (2010) An easy protocol for studying chromatin and recombination protein dynamics during arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129:143–153. https://doi.org/10.1159/000314096
Copenhaver GP, Housworth EA, Stahl FW (2002) crossover interference in arabidopsis. Genetics 160:1631–1639
de Jong JH, Stam P (1985) The association of centromeres of non-homologous chromosomes at meiotic prophase in Beta vulgaris L. Can J Genet Cytol 27:165–171
de Vicente MC, Tanksley SD (1991) Genome-wide reduction in recombination of backcross progeny derived from male versus female gametes in an interspecific cross of tomato. Theor Appl Genet 83:173–178
Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M (2007) Sex-Specific Crossover Distributions and Variations in Interference Level along Arabidopsis thaliana Chromosome 4. Plos Genet 3:e106
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acid Res 7:1869–1885
Grelon M, Gendrot G, Vezon D, Pelletier G (2003) The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J 35(4):465–475
Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M et al (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439:749–752
Harrison CJ, Alvey E, Henderson R (2010) Meiosis in flowering plants and other green organisms. J Exp Bot 61(11):2863–2875
Heribert-Nilsson N (1920) Zuwachsgeschwindigkeit der pollen-schlauche und gestorte Mendelzahlen bei Oenothera lamarchkiana. Hereditas 1:41–67
Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19(20):2488–2500
Higgins JD, Ferdous M, Osman K, Franklin FCH (2011) The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J 65(3):492–502. https://doi.org/10.1111/j.1365-313x.2010.04438.x
Huskins CL, Hearne EM (1933) Meiosis in asynaptic dwarf oats and wheat. J Roy Micr Soc 53:109–177
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. PNAS 101(37):13554–13559
Jauhar PP, Singh U (1969) Desynapsis and the blockage of meiosis in Pennisetum orientale Rich. Theor Appl Genet 39(7):315–319. https://doi.org/10.1007/BF00281912
Ji X (2014) Numerical and structural chromosome aberrations in cauliflower (Brassica oleracea var. botrytis) and Arabidopsis thaliana. PhD thesis, Wageningen University, Wageningen, the Netherlands. ISBN 978-94-6257-160-0
Joyce EF, McKim KS (2011) Meiotic checkpoints and the interchromosomal effect on crossing over in Drosophila females. Fly 5:134–140. https://doi.org/10.4161/fly.5.2.14767
Kaul MLH, Murthy TGK (1985) Mutant genes affecting higher plant meiosis. Theor Appl Genet 70(5):449–466. https://doi.org/10.1007/bf00305977
Klásterská I, Natarajan AT (1975) Stickiness in Rosa meiosis induced by hybridisation. Caryologia 28(1):81–88. https://doi.org/10.1080/00087114.1975.10796599
Klášterská I, Natarajan A, Ramel C (1976) An interpretation of the origin of subchromatid aberrations and chromosome stickiness as a category of chromatid aberrations. Hereditas 83(2):153–162
Koduru PRK, Rao MK (1981) Cytogenetics of synaptic mutants in higher plants. Theor Appl Genet 59(4):197–214. https://doi.org/10.1007/bf00265494
Konvička O, Gottschalk W (1971) Cytologische untersuchungen an strahleninduzierten Mutanten von Brassica oleracea var. capitata. Biol Plant 13(5–6):325–332
Koornneef M, Van der Veen JH (1983) (1983): Trisomics in Arabidopsis thaliana and the location of linkage groups. Genetica 61(1):41–46
Koul KK, Nagpal R (2016) Inter-telomeric connections and achiasmate meiosis in Tradescantia spathacea Sw. Nucleus 59:197–205
Lhuissier FGP, Offenberg HH, Wittich PE et al (2007) The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell Online 19:862–876. https://doi.org/10.1105/tpc.106.049106
Lim KB, de Jong H, Yang TJ, Park JY, Kwon SJ, Kim JS, Lim MH, Kim JA, Jin M, Jin YM, Kim SH, Lim YP, Bang JW, Kim HI, Park BS (2005) Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Mol Cells 19(3):436–444
Lim KB, Yang TJ, Hwang YJ, Kim JS, Park JY, Kwon SJ, Kim J, Choi BS, Lim MH, Jin M, Kim HI, de Jong H, Bancroft I, Lim Y, Park BS (2007) Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J 49:173–183
Mehra RC, Rai KS (1972) Cytogenetic studies of meiotic abnormalities in Collinsia tinctoria II. Desynapsis. Can J Genet Cytol 14:637–644
Menzel MY, Brown MS (1955) Isolating mechanisms in hybrids of Gossypium gossypioides. Am J Bot 42:49–57
Moens PB (1964) A new interpretation of meiotic prophase in Lycopersicon esculentum (tomato). Chromosoma 15:231–242
Pagliarini MS, Freitas PMD, Batista LAR (2000) Chromosome stickiness in meiosis of a Brazilian Paspalum accession. Cytologia 65:289–294
Pedrosa A, Jantsch MF, Moscone EA, Ambros PF, Schweizer D (2001) Characterisation of pericentrometric and sticky intercalary heterochromatin in Ornithogalum longibracteatum (Hyacinthaceae). Chromosoma 110(3):203–213
Peirson BN, Bowling SE, Makaroff CA (1997) A defect in synapsis causes male sterility in a T- DNA-tagged Arabidopsis thaliana mutant. Plant J 11(4):659–669
Rao PN (1975) Desynapsis in Coix lacryma-jobi caused by genotype-environment-interaction. Theor Appl Genet 46(6):315–317. https://doi.org/10.1007/BF00281154
Rayburn AL, Wetzel JB (2002) Flow cytometric analyses of intraplant nuclear DNA content variation induced by sticky chromosomes. Cytometry 49:36–41
Rees H (1961) Genotypic control of chromosome form and behavior. Bot Rev 27(2):288–318
Robinson WP, McFadden DE (2002) Chromosomal genetic disease: numerical aberrations. Encyclopaedia of life sciences. https://doi.org/10.1038/npg.els.0001451
Ross J, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FCH, Jones GH (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA- transformed lines. Chromosome Res 5:551–559
Shi Q, King RW (2005) Nondisjunction, aneuploidy and tetraploidy. Nature 437:1038–1042
Sinha SK, Mohapatra BK (1969) Compensatory chiasma formation in maize. Cytologia 34:523–527. https://doi.org/10.1508/cytologia.34.523
Smyth DR (1995) Flower development. Origin of the cauliflower. Current Biology 5:361–363
Soost RK (1951) Comparative cytology and genetics of asynaptic mutants in Lycopersicon esculentum Mill. Genetics 36:410
Souza AM, Pagliarini MS (1996) Spontaneous chromosome stickiness in canola. Nucleus 39:85–89
Sybenga J (1975) Meiotic configurations. In: Sybenga J (ed) Monographs on theoretical and applied genetics, vol 1. Springer, New York. https://doi.org/10.1007/978-3-642-80960-6
Sybenga J (1992) Cytogenetics in plant breeding. In: Sybenga J (ed) Monographs on theoretical and applied genetics. Springer, New York. https://doi.org/10.1007/978-3-642-84083-8
Sym M, Engebrecht J, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72:365–378
Termolino P, Falque M, Cigliano RA et al (2019) Recombination suppression in heterozygotes for a pericentric inversion induces the interchromosomal effect on crossovers in Arabidopsis. Plant J 100:1163–1175. https://doi.org/10.1111/tpj.14505
Wani AA, Bhat TA (2017) Asynapsis and desynapsis in plants. In: Wani AA, Baht TA (eds) Chromosome structure and aberrations. Springer, New Delhi. https://doi.org/10.1007/978-81-322-3673-3_6
Wei F, Zhang G-S (2010) Meiotically asynapsis-induced aneuploidy in autopolyploid Arabidopsis thaliana. J Plant Res 123(1):87–95. https://doi.org/10.1007/s10265-009-0262-4
Xiong Z, Pires JC (2011) Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187:37–49
Yang M, Nadeau JA, Zhao L, Sack FD (1999) Characterization of a cytokinesis defective (cyd1) mutant of Arabidopsis. J Exp Bot 50(338):1437–1446
Yu H, Wang J, Zhao Z, Sheng X, Shen Y (2019) Construction of a high-density genetic map and identification of loci related to hollow stem trait in Broccoli (Brassica oleracea L. italica). Front Plant Sci 10:45
Zamariola L, De Storme N, Vannerum K, Vandepoele K, Armstrong SJ, Franklin FC, Geelen D (2014) SHUGOSHINs and PATRONUS protect meiotic centromere cohesion in Arabidopsis thaliana. Plant J 77(5):782–794. https://doi.org/10.1111/tpj.12432
Zhao Z, Gu H, Sheng X, Yu H, Wang L, Wang D (2016) Genome-Wide Single-Nucleotide Polymorphisms Discovery and High-Density Genetic Map Construction in Cauliflower Using Specific-Locus Amplified Fragment Sequencing. Front Plant Sci 7:334. https://doi.org/10.3389/fpls.2016.00334
Acknowledgements
The help of Leila Sharifzadeh-Dehkordi MSc in the numerical analyses is appreciated.
Funding
Author X.J. received research support from Rijk Zwaan, R&D Fijnaart, the Netherlands.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by all authors. The first draft of the manuscript was written by Xianwen Ji and Hans de Jong and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ji, X., Lelivelt, C., Wijnker, E. et al. Is partial desynapsis in cauliflower (Brassica oleracea L. var. botrytis) pollen mother cells linked to aneuploidy in the crop?. Euphytica 218, 79 (2022). https://doi.org/10.1007/s10681-022-03027-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03027-7