Abstract
Phenology is one of the most relevant traits for crop development since the duration and timing of plant developmental phases has direct consequences for yield potential. The relative timing of development is even more important in temperate conditions where cool season crops are planted under short days. Our objective was to identify the genomic regions associated with phenology in barley germplasm representative of breeding and production in the Southern Cone of South America. The germplasm was phenotyped for pre-anthesis and post-anthesis phase duration (and photoperiod sensitivity) in nine field experiments evaluated over four years in two locations. The population was genotyped with 1028 SNPs. A comprehensive GWAS was performed and QTL were detected for all traits. Genomic regions were associated with multiple phenology traits in multiple environments. Twenty-five QTL were identified, most of which were associated with more than one trait and/or to the same trait in a different environment. Some of the QTL could be associated with modifications of phase length (PL) or photoperiod sensitivity (PR) and others with both types of response. These findings provide an understanding of phenology in barley in temperate regions and can serve as a starting point for future research into specific genomic regions.


Similar content being viewed by others
Abbreviations
- SE:
-
Seedling emergence
- GDD:
-
Growing degree days
- PL:
-
Phase length
- PR:
-
Photoperiod sensitivity or response
- LD:
-
Linkage disequilibrium
- SEA:
-
Single-environment analysis
- MEMQ:
-
Multi-environment multi-QTL analysis
- ME:
-
Marker effect
- GE:
-
Global effect
- PRA:
-
Phase-reducing allele
- PIA:
-
Phase-increasing allele
References
Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T (2014) Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS ONE 9:e113120. https://doi.org/10.1371/journal.pone.0113120
Alqudah AM, Sallam A, Baenziger PS, Börner A (2020) GWAS: Fast-forwarding gene identification and characterization in temperate cereals: lessons from barley—a review. J Adv Res 22:119–135. https://doi.org/10.1016/j.jare.2019.10.013
Appendino ML, Slafer GA (2003) Earliness per se and its dependence upon temperature in diploid wheat lines differing in the major gene Eps–Am 1 alleles. J Agric Sci 141:149–154. https://doi.org/10.1017/S00221859603003472
Appleyard M, Kirby EJM, Fellowes G (1982) Relationships between the duration of phases in the pre-anthesis life cycle of spring barley. Aust J Agric Res 33:917–925. https://doi.org/10.1071/AR9820917
Arias G, Reiner L, Penger A, Mangstil A (1983) Directory of barley cultivars and lines. Eugen, Stuttgart, p 391
Barret JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. https://doi.org/10.1093/bioinformatics/bth457
Baty F, Ritz C, Charles S, Brutsche M, Flandrois JP, Delignette-Muller ML (2015) A toolbox for nonlinear regression in R: the package nlstools. J Stat Softw 66(5):1–21. https://doi.org/10.18637/jss.v066.i05
Borrás-Gelonch G, Romagosa I, van Eeuwijk F, Slafer GA (2009) Genetic variability in the duration of pre-heading phases and relationships with leaf appearance and tillering dynamics in a barley population. Field Crop Res 113:95–104. https://doi.org/10.1016/j.fcr.2009.03.012
Borrás-Gelonch G, Slafer GA, Casas AM, van Eeuwijk F, Romagosa I (2010) Genetic control of pre-heading phases and other traits related to development in a double-haploid barley (Hordeum vulgare L.) population. Field Crops Res 119:36–47. https://doi.org/10.1016/j.fcr.2010.06.013
Borrás-Gelonch G, Denti M, Thomas WTB, Romagosa I (2012) Genetic control of pre-heading phases in the Steptoe x Morex barley population under different conditions of photoperiod and temperature. Euphytica 183:303–321. https://doi.org/10.1007/s10681-011-0526-7
Boyd WJR, Li CD, Grime CR, Cakir M, Potipibool S, Kaveeta L, Men S, Jalal Kamali MR, Barr AR, Moody DB, Lance RCM, Logue SJ, Raman H, Read BJ (2003) Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L.) genotypes grown over a mild winter growing season. Aust J Agric Res 54:1277–1301. https://doi.org/10.1071/AR03014
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Bullrich L, Appendino ML, Tranquilli G et al (2002) Mapping a thermo-sensitive earliness per se gene on Triticum monococcum chromosome 1Am. Theor Appl Genet 105:585–593. https://doi.org/10.1007/s00122-002-0982-5
Campoli C, von Korff M (2014) Genetic control of reproductive development in temperate cereals. Adv Bot Res 72:131–158. https://doi.org/10.1016/B978-0-12-417162-6.00005-5
Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199:1045–1059. https://doi.org/10.1111/nph.12346
Cantalapiedra CP, Boudiar R, Casas AM, Igartua E, Contreras-Moreira B (2015) BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol Breed 35:13. https://doi.org/10.1007/s11032-015-0253-1
Castro A (2002) Análisis de la coancestría en el germoplasma utilizado en el mejoramiento de cebada en Uruguay. Agrociencia 6:27–39
Castro A, Ernst O, Hoffman E, Bentancur O (1997) Barley germplasm characterization using agronomic and quality traits. Agrociencia 1:80–87
Casao MC, Igartua E, Karsai I, Lasa JM, Gracia MP, Veisz O, Casas AM (2011) Expression analysis of vernalization and day-length response genes in barley (Hordeum vulgare L.) indicates that VRNH2 is a repressor of PPDH2 (HvFT3) under long days. J Exp Bot 62:1939–1949. https://doi.org/10.1093/jxb/erq382
Castro A, Hayes PM, Viega L, Vales I (2008) Transgressive segregation for phenological traits in barley explained by two major QTL alleles. Plant Breed 127:561–568. https://doi.org/10.1111/j.1439-0523.2008.01520.x
Castro AJ, Cuesta-Marcos A, Hayes PM, Locatelli A, Macaulay M et al (2017) The completely additive effects of two barley phenology-related genes (eps2S and sdw1) are explained by specific effects at different periods within the crop growth cycle. Plant Breed 136:663–670. https://doi.org/10.1111/pbr.12509
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A et al (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582. https://doi.org/10.1186/1471-2164-10-582
Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA et al (2007) Control of flowering time in temperate cereals: genes, domestication and sustainable productivity. J Exp Bot 58:1231–1244. https://doi.org/10.1093/jxb/erm042
Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S et al (2012) Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE 7:e45307. https://doi.org/10.1371/journal.pone.0045307
Comadran J, Russell JR, Booth A, Pswarayi A, Ceccarelli S, Grando S et al (2011) Mixed model association scans of multi-environmental trial data reveal major loci controlling yield and yield related traits in Hordeum vulgare in Mediterranean environments. Theor Appl Genet 122:1363–1373. https://doi.org/10.1007/s00122-011-1537-4
Comadran J, Kilian B, RussellJ RL, Stein N, Ganal M, Shaw P et al (2012) Natural variation in a homolog of Antirrhinum centroradialis contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44:1388–1392. https://doi.org/10.1038/ng.2447
Cuesta-Marcos A, Igartua E, Ciudad F, Codesal P, Russell J, Molina-Cano JL et al (2008) Heading date QTL in a spring x winter barley cross evaluated in Mediterranean environments. Mol Breed 21:455–471. https://doi.org/10.1007/s11032-007-9145-3
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184. https://doi.org/10.1016/j.pbi.2008.12.010
Faure S, Higgins J, Turner A, Laurie DA (2007) The Flowering Locus T-like gene family in barley (Hordeum vulgare). Genetics 176:599–609. https://doi.org/10.1534/genetics.106.069500
Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ et al (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109:8328–8333. https://doi.org/10.1073/pnas.1120496109
Federer WT (1961) Augmented designs with one-way elimination of heterogeneity. Biometrics 17:447–473. https://doi.org/10.2307/2527837
Fernández-Calleja M, Casas AM, Igartua E (2021) Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat. Theor Appl Genet 134:1867–1897. https://doi.org/10.1007/s00122-021-03824-z
Ford-Lloyd BV, Newbury HJ, Jackson MT, Virk PS (2001) Genetic basis for co-adaptative gene complexes in rice (Oryza sativa L.) landraces. Heredity 87:530–536. https://doi.org/10.1046/j.1365-2540.2001.00937.x
Gallagher LW, Solliman KM, Vivar H (1991) Interactions among loci conferring photoperiod insensitivity for heading time in spring barley. Crop Sci 31:256–261. https://doi.org/10.2135/cropsci1991.0011183X003100020003x
González FG, Slafer GA, Miralles DJ (2002) Vernalization and photoperiod responses in wheat pre-flowering reproductive phases. Field Crop Res 74:183–195. https://doi.org/10.1016/S0378-4290(01)00210-6
Gutierrez L, Quero G, Fernandez S Brandariz S (2016) lmem.gwaser: linear mixed effects models for genome-wide association studies. R package version 0.1.0. https://cran.r-project.org/src/contrib/Archive/lmem.gwaser/. Accessed 27 July 2021
Gutiérrez L, Germán S, Pereyra S, Hayes PM, Pérez CA, Capettini-Locatelli FA et al (2015) Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor Appl Genet 128:501–519. https://doi.org/10.1007/s00122-014-2448-y
Hay RKM, Ellis RP (1998) The control of flowering in wheat and barley: what recent advances in molecular genetics can reveal. Ann Bot 82:541–554. https://doi.org/10.1006/anbo.1998.0733
Hayes PM, Liu BH, Knapp SJ, Chen F, Jones B, Franckowiak J et al (1993) Quantitative trait locus effects and environmental interaction in a sample of North-American barley germplasm. Theor Appl Genet 87:392–401. https://doi.org/10.1007/BF01184929
He T, Hill CB, Angessa TT, Zhang X, Chen K, Moody D, Telfer P et al (2019) Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection. J Exp Bot 20:5603–5616. https://doi.org/10.1093/jxb/erz332
Hemming MN, Fieg S, James Peacock W et al (2009) Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics 282:107–117. https://doi.org/10.1007/s00438-009-0449-3
Herzig P, Maurer A, Draba V et al (2018) Contrasting genetic regulation of plant development in wild barley grown in two European environments revealed by nested association mapping. J Exp Bot 69:1517–1531. https://doi.org/10.1093/jxb/ery002
Hill WG, Weir BS (1988) Variances, and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78. https://doi.org/10.1016/0040-5809(88)90004-4
Hill CB, Angessa TT, McFawn LA, Wong D, Tibbits J, Zhang XQ (2019) Hybridisation-based target enrichment of phenology genes to dissect the genetic basis of yield and adaptation in barley. Plant Biotechnol J 17:932–944. https://doi.org/10.1111/pbi.13029
Ibrahim A, Harrison M, Meinke H, Fan Y, Johnson P (2018) A regulator of early flowering in barley (Hordeum vulgare L.). PLoS ONE 13(7):e0200722. https://doi.org/10.1371/journal.pone.0200722
Karsai I, Szűcs P, Kőszegi B, Hayes PM, Casas AM, Bedő Z, Veisz O (2008) Effects of photo and thermo cycles on flowering time in barley: a genetical phenomics approach. J Exp Bot 59:2707–2715. https://doi.org/10.1093/jxb/ern131
Kernich GC, Halloran GM, Flood RG (1997) Variation in duration of pre-anthesis phases of development in barley (Hordeum vulgare). Aust J Agric Res 48:59–66. https://doi.org/10.1071/A96020
Kitchen BM, Rasmusson DC (1983) Duration and inheritance of leaf initiation, spike initiation and spike growth in barley. Crop Sci 23:939–943. https://doi.org/10.2135/cropsci1983.0011183X002300050030x
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L) cross. Genome 38:575–585. https://doi.org/10.1139/g95-074
Leon AJ, Lee M, Andrade FH (2001) Quantitative trait loci for growing degree days to flowering and photoperiod response in sunflower (Helianthus annuus L.). Theor Appl Genet 102:497–503. https://doi.org/10.1007/s001220051673
Lewis S, Faricelli ME, Appendino ML et al (2008) The chromosome region including the earliness per se locus Eps-Am 1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot 59:3593–3607. https://doi.org/10.1093/jxb/ern209
Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS (2006) Analysis of QTL for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome 49:454–466. https://doi.org/10.1139/g05-128
Locatelli A, Cuesta-Marcos A, Gutiérrez L, Hayes PM, Smith K, Castro A (2013) Genome-wide association mapping of agronomic traits in relevant barley germplasm in Uruguay. Mol Breed 31:631–654. https://doi.org/10.1007/s11032-012-9820-x
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J et al (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727. https://doi.org/10.1111/tpj.12319
Maurer A, Draba V, Pillen K (2016) Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J Exp Bot 67(8):2507–2518. https://doi.org/10.1093/jxb/erw070
Miralles DJ, Slafer G, Richards R (2000) Duration of stem elongation period influences the number of fertile florets in wheat and barley. Aust J Plant Phys 27:931–940. https://doi.org/10.1071/PP00021
Muñoz-Amatrain M, Moscou ME, Bhat PR, Svensson JT, Bartoš J (2011) An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome 4:238–249. https://doi.org/10.3835/plantgenome2011.08.0023
Obsa BT, Eglinton J, Coventry S, March T, Langridge P, Fleury D (2016) Genetic analysis of development and adaptative traits in three doubled haploid populations of barley (Hordeum vulgare L.). Theor Appl Genet 129:1139–1151. https://doi.org/10.1007/s00122-016-2689-z
Ochagavía H, Prieto P, Zikhali M, Griffiths S, Slafer GA (2019) Earliness per se by temperature interaction on wheat development. Sci Rep 9:2584. https://doi.org/10.1038/s41598-019-39201-6
Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedo Z (1994) Genetic analysis of the components of winterhardiness in barley (Hordeum vulgare L). Theor Appl Genet 89:900–910. https://doi.org/10.1007/BF00224516
Pankin A, Campoli C, Dong X, Kilian B, Sharma R et al (2014) Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198:383–396. https://doi.org/10.1534/genetics.114.165613
Pasam RK, Sharma R (2014) Association mapping: a new paradigm for dissection of complex traits in crops. In: Kavi-Kashor PB, Bandopadhyay R, Suravajhala P (eds) Agricultural bioinformatics. Springer, New Delhi, pp 1–20. https://doi.org/10.1007/978-81-322-1880-7_1
Pham AT, Maurer A, Pillen K et al (2019) Genome-wide association of barley plant growth under drought stress using a nested association mapping population. BMC Plant Biol 19:134. https://doi.org/10.1186/s12870-019-1723-0
Piepho HP, Möhring J (2007) Computing heritability and selection response from unbalanced plant breeding trials. Genetics 177:1881–1888. https://doi.org/10.1534/genetics.107.074229
R Core Team (2018). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doeblay J et al (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484. https://doi.org/10.1073/pnas.201394398
Richards RA (1991) Crop improvement for temperate Australia: future opportunities. Field Crops Res 26:141–169. https://doi.org/10.1016/0378-4290(91)90033-R
Roberts EH, Summerfeld RJ, Cooper JP, Ellis RH (1988) Environmental control of flowering in barley (H. vulgare L.) I. Photoperiod limits to long day responses, photoperiod-insensitivity phases and the effect of low temperature and short day vernalization. Ann Bot 62:127–144. https://doi.org/10.1093/oxfordjournals.aob.a087644
SAS Institute (2004) Statistical analysis system online documentation. SAS, Cary
Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42:899–913. https://doi.org/10.1023/A:1006425619953
Slafer GA (2003) Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann Appl Biol 142:117–128. https://doi.org/10.1111/j.1744-7348.2003.tb00237.x
Slafer GA, Rawson HM (1994) Sensitivity of wheat phasic development to major environmental factors: a re-examination of some assumptions made by physiologists and modellers. Aust J Plant Phys 21:393–426. https://doi.org/10.1071/PP9940393
Snape JW, Butterworth K, Whitechurch E, Worland AJ (2001) Waiting for fine times: genetics of flowering time in wheat. Euphytica 119:185–190. https://doi.org/10.1007/978-94-017-3674-9_7
Szücs P, Blake VC, Bhat PR, Chao S, Close TJ, Cuesta-Marcos A et al (2009) An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2:134–140. https://doi.org/10.3835/plantgenome2008.01.0005
Takahashi R, Yasuda S (1970) Genetics of earliness of growth habit in barley. In: Nilan R (ed) Barley genetics. Washington State University Press, Pullman, pp 388–408
Thomas WTB, Powell W, Waugh R, Chalmers KJ, Barua UM, Jack P et al (1995) Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). Theor Appl Genet 91:1037–1047. https://doi.org/10.1007/BF00223917
Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310:1031–1034. https://doi.org/10.1126/science.1117619
Whitechurch EM, Slafer GA, Miralles DJ (2007) Variability in the duration of stem elongation in wheat and barley genotypes. J Agron Crop Sci 193:138–145. https://doi.org/10.1111/j.1439-037X.2007.00260.x
Worland AJ, Appendino ML, Sayers EJ (1994) The distribution, in European winter wheats, of genes that influence ecoclimatic adaptability whilst determining photoperiodic insensitivity and plant height. Euphytica 80:219–228. https://doi.org/10.1007/BF00039653
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P et al (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644. https://doi.org/10.1126/science.1094305
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A et al (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Aacd Sci USA 103:19581–19586. https://doi.org/10.1073/pnas.0607142103
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20. https://doi.org/10.3835/plantgenome2008.02.0089
Acknowledgements
This research was funded by competitive grants awarded by the Instituto Nacional de Investigación Agropecuaria, Uruguay (FPTA 227), the Fondo Regional de Tecnología Agropecuaria (FONTAGRO, www.fontagro.org) (Project FTG-05617-06), and the Universidad de la República (Programa CSIC Grupos), Uruguay. The authors wish to thank Dr. Patrick M. Hayes (Oregon State University) and Dr. Kevin Smith (University of Minnesota) for their contributions to this research. We also want to express our appreciation for the comments of several anonymous reviewers, which helped to vastly improve our manuscript.
Funding
A funding declaration is mandatory for publication in this journal. Please confirm that this declaration is accurate, or provide an alternative.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10681_2022_2993_MOESM1_ESM.tif
Supplementary Figure 1 Phenotypic distribution of the traits evaluated in each experiment. Below each graph the corresponding statistics are displayed (TIF 6901 kb)
10681_2022_2993_MOESM2_ESM.tif
Supplementary Figure 2 Intra-chromosomal LD (r2) decay of marker pairs over all chromosomes as a function of genetic distance (cM). The horizontal line (red dotted line) indicates the 95th percentile distribution of unliked r2). The fitting curve (green line) indicates the LD decay. The vertical line (blue dotted line) indicates the genetic distance value (2.54) when the fitting curve cross the critical r2 value (0.2) (TIF 1223 kb)
10681_2022_2993_MOESM3_ESM.tif
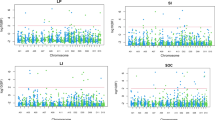
Supplementary Figure 3 Circular-Manhattan plot of [−log 10 (P-values)] vs chromosomal position of SNP markers associated with each of the phenological traits evaluated, either as LP (a) or PR (b). Black-dashed line represents the association threshold p < 0.005; (−log10 (P-values) ? 2.3. The scale (−log10 (P-values) is shown from the center to the top of the figures. Each marker-trait association above the threshold is represented by red points. The most important markers detected by SEA and MEMQ are highlighted. The rings represent the seven phenological traits evaluated (TIF 16085 kb)
10681_2022_2993_MOESM4_ESM.tif
Supplementary Figure 4. Correlations matrix among traits evaluated in each experiment. The distribution and H2 of each trait are shown on the diagonal. GDD for each phenological phase are displayed on the axes. Broad sense heritability estimates (displayed on the bottom of each histogram) were calculated following Piepho and Möhring 2007. On the bottom of the diagonal the bivariate scatter plot with a fitted line was presented. On the top of the diagonal the correlation value (Pearson method) plus the significance level as stars are shown. Each significance level is associated with a symbol: p-values (0.001, 0.01, 0.05, 0.1, 1) (“***”, “**”, “*”, “.”, “ ”) (TIF 16008 kb)
10681_2022_2993_MOESM5_ESM.tif
Supplementary Figure 5 Pairwise Spearman correlation among experiments for Z20-Z30, Z30-Z49, SE-Z49 and Z49-Z90 generated with corrplot package (version 0.88) in R, using corrplot function. The colors represent the correlation sign (blue and red, positive and negative correlation, respectively) (* P <0.01; ** P <0.001; *** P <0.0001) (TIF 1467 kb)
10681_2022_2993_MOESM6_ESM.tiff
Supplementary Figure 6 Principal component analysis of the population using all informative SNPs, highlighting the genotypes involved in the LD among chromosomes. In the main LD complex, the genotypes with allele "0" for the markers; 11_20032 (2H), 11_20130 (3H), 11_10177 (5H), 11_20230 and 11_20103 (7H) were grouped together in the PCA analysis (red points). In the other LD complex, the genotypes with allele "0" for markers; 1120838 and 1120053 were also grouped together in the PCA analysis (blue points). The rest of the genotypes had other allelic combinations for the mentioned markers (grey points) (TIFF 894 kb)
Rights and permissions
About this article
Cite this article
Locatelli, A., Bhatta, M., Gutiérrez, L. et al. Genetic control of barley phenology in South American environments. Euphytica 218, 53 (2022). https://doi.org/10.1007/s10681-022-02993-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-02993-2