Abstract
Quantitative trait loci for yield, yield components and seed protein content were investigated on the basis of experiments performed with two populations of pea (Pisum sativum L.) lines derived from linked crosses between lines Wt11238, Wt3557 and Wt10245 with contrasting characteristics. The yield-related traits were defined as components giving the grain yield in a multiplicative way. The aim was to clarify the genetic architecture of the relation between seed yield, its components and protein content, with a possible inclusion of the role of epistasis in this explanation. To take full advantage of the availability of the two populations, additive QTL effects and both types of epistasis were analysed: the QTL by genetic background interaction and the first-order QTL–QTL interaction. The two hybrid populations differed with respect to the prevailing gene action, which in the Wt11238 × Wt3557 progeny was mainly additive, while in the Wt10245 × Wt11238 progeny mainly epistatic. Some loci with previously reported, large, repeatable, but contradictory effects on yield and protein content were confirmed. New loci with alleles coming from the protein-rich Wt11238 line, positive for yield components, were identified. It was found that the first order QTL–QTL interaction events were more frequent for the loci showing the QTL by genetic background interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pea (Pisum sativum L.) is considered as one of the most important legume crops in Europe. Unfortunately, even its cultivars can perform in an unstable way across different environmental conditions (years and locations), as discussed e.g. by Timmerman-Vaughan et al. (2005). The instability may concern grain yield or its components, which show an increasing environmental variation according to the order: number of seeds per pod, number of pods per peduncle, number of fertile nodes per plant, number of pods per plant, and number of seeds per plant. Moreover, a mutual compensation of yield components has been known (Moot and McNeil 1995). Pea cultivars are also expected to provide high amounts of protein. As negative correlations between seed yield and protein content were reported for many crop plants including pea, the two requirements may be contradictory. Święcicki et al. (1981) described progress in breeding of cultivars with higher protein contents and high yield as less significant than expected.
Today, localization of quantitative trait loci (QTL) has become a standard approach for finding genomic regions responsible for agriculturally important traits. Although pea was once used as a model plant for studying inheritance of qualitative and quantitative traits, not many reports concern its QTLs. Timmerman-Vaughan et al. (1996) mapped QTLs conditioning seed weight using two populations, the progeny of a cross between two cultivated types (Primo and OSU442-15) and single-seed-descent recombinant inbred lines from a wide cross between a P. sativum subsp. sativum line (Slow) and a P. sativum subsp. humile accession (JI1794). Eight loci were found in both linkage maps. In the two crosses, only one common genomic region was identified as containing the seed-weight QTL. Tar’an et al. (2004) conducted the experiment to identify loci responsible for grain yield, seed protein content and maturity. One population of 88 recombinant inbred lines developed from a cross between Carneval and MP 1401 was observed in 13 environments in three vegetative periods. Eleven loci were found for all observed traits. Irzykowska and Wolko (2004) constructed a linkage map of pea and used it for interval mapping of QTLs controlling seed number per plant, pod number per plant, 1000-seed weight, seed yield, and seed protein content. The yield-related traits were measured in generations F2 and F4. A total of 37 QTLs were detected for all traits. Timmerman-Vaughan et al. (2005) analyzed loci for seed yield, yield components and developmental traits. One mapping population, of F2-derived families from the cross between Primo × OSU442-15, was used. The experiment was repeated three times. Nineteen loci were found for all traits.
Despite some success of QTL studies in crop plants, several of them show poor reproducibility of QTL locations and effects over different material obtained for the same species. It is indicated that a primary reason for this should be seen in the possibility of a non-allelic interaction (epistasis). Some insight into this problem can be gained by considering the linked biparental crosses. A possibility to formally study epistasis in linked crosses was noticed by Charcosset et al. (1994), who considered a contrast of additive effects estimated in three linked crosses as a measure of interaction. Later, Jannink and Jansen (2001) considered a similar contrast approach to build linear models and a likelihood method to test for the presence of epistasis. More recently, Blanc et al. (2006) performed a study of QTLs in six populations obtained in a diallel design. In the papers cited here (as well as others), two types of epistasis are discussed (Carlborg and Haley 2004): the QTL by genetic background interaction and the QTL–QTL first order interaction. The former can be studied only when several populations are investigated which provide a spectrum of backgrounds. The latter can be measured also in a single population, using one of several algorithms available in the literature and in the QTL analysis software.
The aim of the study reported here was to investigate QTLs for yield, yield components and protein content, on the basis of multi-environment trials performed with two populations of pea lines, with the hope to clarify the genetic architecture of the relation between seed yield, its components and protein content, and with a possible inclusion of the role of epistasis in this explanation. The study involved two populations of progeny derived from two linked crosses between lines Wt11238, Wt3557 and Wt10245 with contrasting characteristics, observed in four seasons and at two locations. Unlike in the other reports, the yield-related traits were defined as components giving grain yield in a multiplicative way. Full advantage was taken of the availability of the two populations. Besides additive QTL effects, found by the application of Windows QTL Cartographer 2.0 (2007) software, both types of epistasis were analysed: the QTL by genetic background interaction by a comparison of additive QTL effects found, and the first-order QTL–QTL interaction by the recently developed inclusive composite interval mapping (ICIM) method (Li et al. 2008).
Materials and methods
Plant material and field experiments
Two populations of F2-derived lines were obtained from the crosses: Wt11238 × Wt3557 (population Q1, 110 lines of the F5 generation in the first year of the experiment) and Wt10245 × Wt11238 (Q2, 101 lines of the F5 generation; earlier generations of this material were used by Irzykowska and Wolko 2004). The parental lines were selected on the basis of contrasting features: Wt3557 and Wt10245 are large-seeded, high-yielding cultivars, while Wt11238 is a small-seeded, low-yielding, but protein-reach line, marked by interesting mutations in several morphological loci (d, tl, r, cp, wb, s, i, u, te, gp; see Święcicki 1987). The populations of hybrids were obtained by selfing for several generations (F3 seeds representing individual F2 plants were harvested; at least five F3 plants were grown to produce F4 seeds, and the resulting seeds were bulked; the cycle was repeated for subsequent generations). Populations were observed in the field (in randomized blocks, in 2 or 3 replications) in the years 2003–2005 at Poznań (52°26′44′′ N, 16°54′04′′ E) and Wiatrowo (52°45′18′′ N 17°08′19′′ E) (5 plants per plot). Five phenotypic traits of plants were observed in this design: peduncle number (PED), pod number per peduncle (POD), seed number per pod (SEED), thousand seed weight (in g, TSW) and yield (in g, YIELD). The sixth trait, seed protein content (Kjeldahl method using Kjel-Foss nitrogen analyzer, %, PROT), was observed four times in each population (2002 in Poznań, 2003 in Poznań and Wiatrowo, and 2004 in Wiatrowo). Fertilizers, insecticides and herbicides were applied as required and in accordance with local practices.
Genotyping and linkage analysis
Observations of morphological, isozymic and molecular (AFLP, RAPD, STS, CAPS, ISSR) marker polymorphisms were performed in earlier projects. Individual F2 plants were scored for morphological markers and several isozyme markers. F4 lines (bulks of 5 plants) were scored for the rest of isozyme markers and DNA markers. Details of AFLP markers are presented by Irzykowska et al. (2001) and Gawłowska et al. (2005). For the present analysis the genetic characterization of lines was supplemented by observations of a number of SSR markers taken from the model legume Medicago truncatula L. Three polymorphic SSR markers (MTIC 4 [TC 28977], MTIC 153 [TC 36422], MTIC 451 [TC 32263]) were located in both maps in linkage groups (LG) VI and I. MTIC 16 [TC28470] was mapped in LG IIIb in the Q2 map only, and MTIC 223 [TC 35288] was placed in the additional group VIII in the Q1 map. The linkage maps, constructed by the maximum likelihood method in JoinMap ver. 3.0. (Van Ooijen and Voorrips 2001), are characterized in Table 1.
Statistical analysis
Analysis of variance with location effects and lines × location interaction effects was conducted independently for each year (with the exception of PROT in 2002 and 2004, where only effects of lines were considered). Correlations between traits were calculated on the basis of mean values for lines in all experiments. Repeatability of results over generations was estimated by correlation coefficients of mean values for lines observed in consecutive years. These computations were done in Genstat (VSN International Ltd. 2010).
For localization of QTLs with additive and dominance effects, the Windows QTL Cartographer 2.0 (2007) software was used for composite interval mapping (CIM), independently for each trait and each experiment (option cross SFn, where n = 4, 5, 6, 7 for data from the consecutive years). Using the statistics reported by the program, at each interval mapping step first the hypothesis concerning the additive QTL effect was tested. Then, depending on the result of this test, the hypothesis concerning the dominance effect was tested in the model with or without the additive effect. The additive and dominance effect estimates were taken from the accepted models. QTLs exhibiting significant effects (LOD > 3) in more than one experiment (in map positions close to the same or directly neighbouring markers) were selected for interpretation.
The ICIM method (Li et al. 2008) was used to find first-order QTL–QTL interaction effects. As the method is restricted to the analysis of homozygous lines, heterozygotes were treated as missing genotypes for codominant markers and as dominant homozygotes otherwise. Interactions repeated in at least two experiments (LOD > 3), concerning pairs of QTLs separated by more than 20 cM, were selected.
Results
Trait characteristics
Mean values for all observed traits in all experiments are shown in Fig. 1a, b. Populations Q1 and Q2 reacted to variable environmental conditions in a similar way. TSW was always larger in Q2. PROT was always greater in Q1, but the differences were not substantial taking into account the estimated standard errors. When averaged over all experiments, PED and SEED were larger in Q1, TSW in Q2, whereas POD, YIELD and PROT were approximately equal for the two populations. For yield-related traits, the largest variability of the means between years was observed for PED and YIELD, intermediate for POD and SEED, and the smallest for TSW. The differences between locations within years were highly significant in a majority of cases (except for PED in Q1, 2004; POD in Q2, 2004; TSW in Q2, 2003 and 2005; YIELD in Q1, 2005). Values of SEED observed within years were always significantly smaller in Poznań than in Wiatrowo; the same was almost always true for TSW and YIELD. Thus, the mean values in the two locations (over both populations and 3 years) for SEED and TSW were substantially smaller in Poznań, which resulted in a much smaller average YIELD in Poznań (8.57 g) than in Wiatrowo (13.01 g), whereas the corresponding mean values for PED and POD were similar. The mean value for PROT was larger in Poznań (28.19%) than in Wiatrowo (26.86%).
a Mean values of the yield-related traits for Q1 and Q2 in all experiments. b Mean values of the protein content for Q1 and Q2 in all experiments. c Optimum monthly precipitation and monthly precipitation observed in the locations of the experiments. “Po” and “Wi” denote experiments performed in Poznań and Wiatrowo, respectively
The monthly mean air temperatures and total precipitation observed between March and July at the two locations in 2003–2005 were used in an attempt to explain the variability of mean values for yield components. No role of monthly temperatures could be proved. It was observed that the profiles of mean values for PED and YIELD, and to some extent for POD, show a correspondence with the profiles of precipitation values for May, June and July (Fig. 1c), which differed between years, and also between locations, especially in 2005. More precisely, a significant negative correlation was found between PED and absolute deviations of monthly precipitations from the levels found as optimal for pea by Święcicki and Święcicki (1981): 65, 70 and 45 mm, respectively, in May, June, and July (r = −0.701, −0.678, −0.782; P < 0.01); a negative correlation was found between POD and the deviations for precipitation in June (r = −0.654; P < 0.05). This analysis explains to a great extent why high PED values were observed in 2004 (15.58–21.21), when the precipitation was stable over the vegetation period and close to optimum, and low in 2003 (5.69–11.28) and in 2005 (6.82–12.34), when the precipitation in July and May, respectively, was larger than optimum. Large PED in 2004 in Wiatrowo created the highest YIELD (18.73–20.34 g), whereas YIELD in Poznań was reduced by relatively small values of SEED and TSW. The lowest values of PED in 2003 in Poznań caused the smallest YIELD (4.15–4.51 g). No strict relation between weather conditions and protein content could be observed; the highest PROT level (29.00–29.25%) was reached in 2003 in Poznań, when YIELD was the lowest.
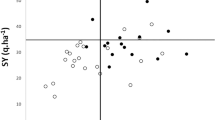
The range of mean values for lines, calculated over all experiments, was usually larger than the difference between mean values for the parental lines, with the exception of TSW in Q2 (Fig. 2). The behaviour of parental lines was in accordance with their known properties: Wt3557 was better than Wt11238 with respect to all yield-related traits; Wt10245 was better than Wt11238 with respect to all yield-related traits except for SEED. Lines Wt3557 and Wt10245 were approximately equivalent for PED, POD, YIELD and PROT. However, they differed greatly with respect to SEED, with Wt10245 being inferior to the two other parental lines, and with respect to TSW, with Wt10245 being superior to the two other parental lines. Wt11238 was markedly superior to both Wt3557 and Wt10245 for protein content.
Analysis of variance for yield and yield components showed that the pattern of occurrence of the interaction of lines with locations was similar in populations Q1 and Q2 (Table 2). It differed only for PED in 2004 and YIELD in 2003, where the interaction was significant in Q2, but not in Q1. The interaction was always significant for SEED, and in two out of 3 years for TSW. In 2005, interaction of lines with locations was significant in both Q1 and Q2 for all yield-related traits. For PROT, the interaction of lines and locations could be measured only in 2003, and was non-significant both in Q1 and Q2.
The repeatability of observations in the consecutive generations, measured by the correlation coefficients of mean values for lines, was similar when estimated for F6–F7 and F7–F8; the mean correlations are shown in Table 2. The correlation was statistically significant in all cases except for POD in Q2; it was also markedly smaller for PED in Q2 than for the other traits. The highest mean correlations were observed for TSW and PROT in Q1, and for TSW and SEED in Q2.
The pattern of statistically significant correlations between observed traits is shown in Fig. 3. In Q1, PED was correlated with POD, and SEED with TSW. In Q2, the yield components were not correlated. In both Q1 and Q2 all components were significantly correlated with YIELD. Seed protein content was negatively correlated with YIELD.
QTL localization
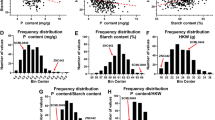
QTL mapping by the CIM method located, in total, 77 significant additive QTL effects for all analyzed traits in Q1 (49 effects) and Q2 (28). For yield-related traits, the number of effects observed in 2003, 2004 and 2005 was 29, 19 and 16, respectively, while for Poznań and Wiatrowo it was 27 and 37, respectively. The greatest number of effects was found for Wiatrowo, 2003 (21), and the smallest for Wiatrowo, 2005 (5). For PROT the distribution of QTLs was almost uniform over four experiments (4, 4, 2, 3). For different traits the number of QTL effects was from 6 for YIELD to 12 for TSW in Q1, and from 4 for YIELD to 7 for SEED and TSW in Q2.
Out of all the 77 additive effects the number of effects repeated in at least two experiments was nine in Q1 and five in Q2. They are summarized in Table 3a, b and in Fig. 4a, b, and described below.
In Q1, mapping provided QTLs in linkage groups I, II, IIIa, Va and Vb (Table 3a). Wt3557 provided positive QTL alleles for POD, TSW and YIELD, whereas Wt11238 for PED, SEED and PROT. Two regions containing QTLs related to more than one trait were observed: region A in LG IIIa, containing markers C10a and a45q, was found to be related to PED and TSW; region B in LG Vb (cp, gp, te)—to TSW, YIELD and PROT.
In Q2 mapping provided QTL regions in linkage groups I and II. Wt10245 provided positive QTL alleles for PED, TSW and YIELD, and Wt11238—for SEED.
Taking into account mapping results in both Q1 and Q2, the region X common to the two maps, around the common markers afp15h and a, was found to be related to SEED.
In addition to additive effects reported above, the applied interval mapping procedure allowed us to identify regions with significant dominance effects of QTLs. No such regions repeated in more than one experiment were found in Q1. In Q2 nine regions related were found. Here we report the only region repeated three times—containing markers (afp1h, Pl), found to be related to YIELD, with the average dominance effect estimated at 12.49 (mean LOD = 5.88).
First-order QTL–QTL interactions
Results of the analysis of first-order QTL–QTL interaction effects by the ICIM method are summarized in Table 4. In one case out of three concerning SEED in Q2, the region involved in interactions was declared as containing also a marker linked closely to an additive QTL for the same trait (afp15h in LG II).
QTL by genetic background interaction
Table 5 contains a summary of additive QTL effects (LOD > 2) and first-order QTL–QTL interaction effects (LOD > 3) found in the ±5 cM neighbourhood of 24 markers, which were common to the linkage maps of Q1 and Q2 (indicated by dots in Fig. 4).
The additive effects of QTLs located in the neighbourhood of common markers were averaged for each marker independently, giving an estimate of the mean additive effect of marker regions in Q1 and in Q2 (results not shown). Then, for each trait, all 24 common markers were classified into one of the two categories (Table 5): (1) markers with additive effects absent both in Q1 and in Q2 or present both in Q1 and Q2, (2) markers with an additive effect present either in Q1 or in Q2. Category (1) represents agreement of additive effects in the two populations, i.e., a situation in which there is no QTL by genetic background interaction, whereas category (2) reflects some interaction of a QTL with the genetic background. The number of cases in categories “1” and “2” was 124 and 20, respectively (in category “1” the case of additive effects present in both Q1 and Q2 occurred just once and the signs of the effects were the same). Further, the traits were characterized by the mean total (in Q1 plus in Q2) number of first-order QTL–QTL interaction effects in the neighbourhood of common markers, with a subdivision of the markers into the two categories described above. The obtained values were close or larger in category “QTL by genetic background interaction present” for POD, SEED, TSW and YIELD, and when averaged over all traits (0.13 vs. 0.35). The comparison was not possible for PED, where no markers showing QTLs by background interaction were present.
To prevent an incorrect interpretation of the QTL by genetic background interaction we calculated the proportions of cases showing a QTL by background interaction for the group of morphological, izosymic and STS markers, for which the two parental lines Wt3557 and Wt10245 could be recognized as having the same alelle, and for the group of SSR, AFLP and CAPS markers, for which this information was not obtained. The two proportions were 0.21 and 0.07, respectively (χ2 = 5.81, 1 df, P = 0.016). Therefore, it cannot be said that the observed interaction is caused by the fact that for SSR, AFLP or CAPS markers there is a possibility that the two parental lines differentiating the two crosses have different marker alleles.
Discussion
Phenotypic relationships
The multi-environment experiments described in this paper provided a picture of genetic architecture of yield-related traits and of total protein content in the two studied populations of pea lines. In terms of general mean values of the traits the two populations reacted similarly to variable conditions represented in 3 years and at two locations. The conditions recorded in the Wiatrowo trial, located in a rural location in Northern Wielkopolska, were more favourable than those in the Poznań trial, located in the suburban area of a large city, both for Q1 and Q2, in terms of SEED and TSW, which can be linked to results showing a dependence of pea physiology and of certain yield components on light intensity and CO2 content in the air (Wager 1974; Hole and Hardwick 1976).
Weather conditions differed considerably in the 3 years. It is known that the precipitation level is important for the development of pea plants: insufficient watering in May can cause problems with flowering and pod setting, and excessive rain in July causes non-uniform maturity (Święcicki and Święcicki 1981). In our experiments the most favourable conditions occurred in 2004. In that year precipitation was distributed evenly over May, June and July, and was relatively close to values considered as optimal for pea (Święcicki and Święcicki 1981), which was found to be related to good plant development, especially in terms of PED, and to some extent—POD. In 2003 and 2005 precipitation was excessive in July and May. The observed relation between precipitation and quality of development agrees with the results of Ney et al. (1994), who applied drought at different developmental stages of pea and concluded that insufficient watering at early flowering reduces the number of flowering nodes. Maurer et al. (1968) claimed that excessive precipitation before and after flowering results in unfavourable changes in plant architecture.
The relations between mean values of the traits for populations Q1 and Q2, calculated over all experiments, reflected to a large extent the relative properties of parental lines Wt3557 and Wt10245, which had been known beforehand, decided on their usage in the study, and were confirmed in the field. The superiority of Wt3557 in terms of SEED resulted in larger values of this trait in Q1, while the superiority of Wt10245 in terms of TSW resulted in larger values of that trait in Q2. For POD both the two parental lines, Wt3557 and Wt10245, and their progeny resulting from crossing both with Wt11238, were equivalent. Only for PED the general rule seemed to be disturbed, as Wt3557 and Wt10245 were on average similar for that trait, but Q1 had on average larger values than Q2. This relation was, however, not the same in all experiments. The final effect, YIELD, was similar in both Wt3557 and Wt10245, and in Q1 and Q2, although the way in which it was reached is clearly different in Q1 (through relatively higher SEED, inherited from Wt3557) and in Q2 (through relatively higher TSW, inherited from Wt10245). Q1 and Q2 were also, on average, equivalent for PROT.
The behaviour of individual pea lines in the experiments can be discussed in relation to two factors. The interaction of lines with locations within years was purely environmental and its pattern was similar for both sets of lines. The fact that it occurred to a bigger extent in 2005 can be putatively ascribed to the largest difference in precipitation distribution between Poznań and Wiatrowo in that year. On the other hand, the differential reaction of lines in consecutive years, which was caused both by different weather conditions and imperfect repeatability of the traits over generations, was not quite the same in Q1 and Q2, especially for PED and POD. By relating this fact to the interactions observed within years we can attribute that difference to a smaller broad sense heritability of those traits in Q2, also in relation to other traits in the same population.
The way in which the phenotypic traits were recorded facilitated the dissection of yield into four components: peduncle number, pod number per peduncle, seed number per pod, and thousand seed weight, which turned out to be weakly correlated with one another. Only in Q1 were significant, positive correlations observed between PED and POD, and between SEED and TSW. All considered yield components were significantly correlated with yield, with the highest correlation observed for peduncle number. Analysis of trait mean values in the experiments also revealed that a high peduncle number was the main yield-creating factor. Discussion of our results with the ones obtained by other authors is difficult, as the reports differ with respect to the definition of observed traits. The negative correlation between the number of pods per plant and the number of seeds per pod was described by Moot and McNeil (1995). According to Poggio et al. (2005), seed number per square meter was the dominant determinant of the variation of pea yield across sowing dates and within cultivars of similar seed weight. A higher number of seeds per square meter can be compensated by a lower seed thousand weight, especially in large-seeded cultivars, in which assimilate production may be insufficient to cover the demand of seed development. As to the correlation between yield (and yield components, correlations not shown) and total protein content, it was negative, which agrees with earlier reports of Święcicki et al. (1981) and Bertholdsson (1990).
Individual QTL effects
Despite the environmental variation, the significant additive QTL effects that were found were evenly distributed over years and locations. Markedly smaller numbers of loci were found for PED and POD, the traits with lower heritability. More repeated additive effects were found in Q1 than in Q2. One of the additive effects found for yield components in Q1 was reflected by a QTL for yield itself, but no such case was observed in Q2. Pleiotropic effects on yield components were observed, but it is our opinion that pleiotropism resulting from statistical correlations between traits was removed due to the applied definition of the traits. One of the QTLs found for PROT had a corresponding QTL for yield-related traits. A comparison of the found QTL regions with other published results is complicated due to different linkage maps used and to different definitions of the observed yield components; however, some overlapping could be found and is discussed in details below.
In LG V an important region was found around markers cp, gp and te. It contained QTLs for TSW, YIELD and PROT in Q1 (region B). The alleles of Wt11238 had a negative influence on yield components and positive on protein content. This may have been expected, since cp, te and gp, respectively, mark concave, narrow and yellow pods of Wt11238, being less suitable for developing a high yield than convex, broad and green pods of the two other parental lines, due to their smaller photosynthetic potential (Flinn and Pate 1970). Timmerman-Vaughan et al. (1996) detected a locus for TSW in LG V near marker P445; according to Weeden et al. (1996, 1998) P445 is closely linked to a viciline Vc-2 gene, which in turn lies very close to cp, approximately 11 cM from gp and 3 cM from te. Irzykowska and Wolko (2004) also reported a location of a TSW locus in LG V close to te.
A region around tl (and r) was found to be linked to PROT in Q1, with the positive alleles from Wt11238, similarly as in Irzykowska and Wolko (2004). Such effects of the tl and, especially, r genes, whose mutant alleles cause, respectively, “acacia” type leaves and wrinkled seeds, are known to breeders and reported, e.g., by Shia and Slinkard (1977) and Davies (1980). Although both Q1 and Q2 lines segregated with respect to those genes, a significant positive additive effect of r mutation on PROT in Q2 was declared only in one experiment, which should be attributed to the generally lower heritability in Q2 than in Q1.
A region around marker afp15h was found to be linked to SEED in Q1 and in Q2 (region X). Marker afp15h is approximately 18 cM from the LegK marker. Fuchs et al. (1998) assigned the legumin K locus to chromosome 6, identified here as linkage group II. A cluster of genes encoding legumins J, K, L, and M was also mapped genetically in linkage group II.
A region around marker afp9i was found to be linked to YIELD in Q2. Marker afp9i is located 13 cM from the k gene, which was found by Matta and Gatehouse (1982) to be linked (approx. 3 cM) to a gene coding convicilin, a storage protein (Cvc).
One should note that our results show a restricted similarity to the ones of Tar’an et al. (2004), who detected four QTLs for yield, one of them in LG II, similarly as our region in Q2. However, a precise comparison of these locations is not possible because of a lack of common markers in both maps. Tar’an et al. (2004) identified also three genomic regions that explained a large portion of the total phenotypic variation for seed protein content. Their localization shows no correlation with ours, because their QTL regions occupy LGs III, VI and A. Finally, the same authors reported no QTL regions affecting simultaneously yield level and protein content. In contrast, our results (region B) seem to have a better correspondence with the observed negative correlation between seed yield and protein content and are likely effects of some genetic basis of that correlation.
The QTLs discussed above have large and repeatable effects, but their linkage to protein content QTL or to legumin and convicilin genes imply that they may have an unfavorable feature of affecting both yield-related traits and protein content in a contradictory manner. They are of limited practical importance, even though, according to Święcicki et al. (1981), the negative protein-yield correlation is strong only above the protein level of about 27%. However, our study allowed us to find some QTL regions with better properties. Several loci with Wt3557 and Wt10245 alleles, positive just for yield components, were found, but this was expected due to the known value of these lines. It is more interesting to enumerate the regions carrying Wt11238 alleles positive for yield components, but without a negative effect on PROT. Wt11238 was found to carry positive alleles in region “A” around C10a and a45q (in Q1) for PED, with a compensating negative effect on TSW, and for SEED in a QTL linked to afp15h and a in Q1 and Q2. Moreover, a positive effect on PROT only of Wt11238 alleles was found close to tl in LG Va. These QTL regions could be further studied for an application in creating genotypes with a high seed protein content and a high seed yield.
Dominance effects estimated in QTL Cartographer were clearly overestimated. The reported mean dominance effect was about 12, whereas the largest additive effect found for yield did not exceed 2.5. The value of 12 is also inconsistent with the estimates of the mean yield for lines. Therefore, it seems that the applied genetic model of segregation in F2-derived lines is able to indicate the situations, in which dominance may be present, but a precise estimation of the parameters is not feasible. A possible source of this may lie in the restricted population sizes or small plots used in the experiments, in which the expected fractions of homo- and heterozygous plants for non-homozygous lines might not be correctly represented, or affected by a plot-to-plot variability. To obtain further explanation of this situation computations were made using our own scripts, which used the segregation model analogous to the one applied in QTL Cartographer, but assuming that all QTLs are located at marker loci. The obtained results coincided in most cases with the ones reported in this paper with respect to the magnitude and the sign of additive parameter estimates. However, it was found that overestimation of dominance effects might be removed by assuming that the fraction of heterozygotes among the plants representing individual lines was larger than that predicted by the probabilistic considerations, which actually may be true in the populations studied.
Epistasis
Besides additive QTL effects, our study allowed us to obtain information about the epistatic gene action by a model-based analysis of first-order QTL–QTL interaction effects in the two studied populations and by a comparative analysis of additive QTLs found. Timmerman-Vaughan et al. (2004) used a similar experimental approach of two linked crosses to study QTLs for blight resistance, but without a formal analysis of epistasis. Our material was not perfect for studying the QTL–QTL interaction and QTL by genetic background interaction for several reasons. Firstly, only two linked crosses were available. Secondly, the maps were quite different with respect to the number of markers and the marker density. Finally, the number of common markers was low. Thus, only an approximate analysis of the epistasis could be done.
Our comparison of two hybrid populations showed that although their average reaction to changing environmental conditions was quite similar and their average seed and protein yields were close, the gene action was quite different. This can probably be partially explained by the fact that line Wt11238 acted as the paternal line in one of the crosses and as the maternal one in the other. Yield components were more inheritable in Q1, with additive gene effects dominating the nonallelic interaction effects. In Q2 more interaction effects were found than additive ones and at least two yield components, PED and POD, had a limited heritability. On the other hand, the QTL region that was significant in both Q1 and Q2 for the same trait, i.e. X for SEED, showed a consistency of the positive effects of the alleles coming from Wt11238.
The results of the analysis of first-order QTL–QTL interaction effects in some cases could yield additional information about the additive QTLs found. It shows that besides a QTL in region B in Q1 with an effect on POD, the same region could carry a QTL for PROT with an effect conditioned by a locus linked to Lap1 in LG IIIb. The significant interaction of the afp15h locus for SEED in Q2 limits the potential of the Wt10245 alleles in that locus.
Our analysis of the QTL by genetic background interaction was based on the estimation of the (average) additive QTL effects found in the vicinity of the 24 markers common for the two linkage maps. We checked that the observed interaction was not an artifact caused by an improper recognition of parental alleles for some of the markers. Then we tried to explain their consistency or inconsistency in Q1 and Q2 by the number of QTL–QTL interaction effects found in the corresponding regions. The mean number of first-order QTL–QTL effects was almost three times larger for the regions showing inconsistency of additive effects than for the regions with consistent effects, which shows some positive relationship between the applied measure of the QTL by genetic background interaction and the measure of the pairwise QTL–QTL interaction. This relation was not consistently such for all traits, so although some correspondence between the two types of non-allelic interaction was proved, it is not universal, as already noted by Blanc et al. (2006).
One should note that our considerations of the QTL by genetic background interaction correspond in some sense to the idea of using a statistical model based on the contrast of additive effects estimated in three linked crosses, as considered by Charcosset et al. (1994) and Jannink and Jansen (2001). The difference is that here, having just two linked crosses, we assume that the additive affect in the third (not available) cross is zero (or no segregation). Our measure of interaction is a categorical one and assigns “no interaction” to all cases with the contrast close to zero and “interaction” to all cases with a non-zero contrast. Our experiments indicated no regions with significant, but contradictory (different sign) additive effects in the two populations. All regions assigned to the category showing some QTL by genetic background interaction were characterized by a significant additive effect in one of the two populations and by a null effect in the other. The actual values of the contrast calculated for all traits and all common markers showed a weakly significant rank correlation with the number of QTL–QTL interaction effects (P < 0.1; data not shown). This adds some strength to our claim that it may be true that the first order QTL–QTL interaction events are more frequent for the loci showing the QTL by background interaction. In any case, it is our opinion that for studies with more linked crosses and more evenly saturated linkage maps with numerous common markers, the described method of analysis may be used as an exploratory alternative to the model-based analysis described by Blanc et al. (2006) or Jannink and Jansen (2001).
References
Bertholdsson NO (1990) The influence of the pea plant ideotype on seed protein content and seed yield. J Agron Crop Sci 164:54–67
Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L (2006) Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet 113:206–224
Carlborg Ö, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5:618–625
Charcosset A, Causse M, Moreau L, Gallais A (1994) Investigation into the effect of genetic background on QTL expression using three connected maize recombinant inbred lines (RIL) populations. In: Ooijen JWV, Jansen J (eds) Biometrics in plant breeding: applications of molecular markers. CPRO-DLO, Wageningen, pp 75–84
Davies DR (1980) The ra locus and legumin synthesis in Pisum sativum. Biochem Genet 18:1207–1219
Flinn AM, Pate JS (1970) A quantitative study of carbon transfer from pod and subtending leaf to the ripening seeds of the field pea (Pisum avense L.). J Exp Bot 21:71–82
Fuchs J, Kühne M, Schubert I (1998) Assignment of linkage groups to pea chromosomes after karyotyping and gene mapping by fluorescent in situ hybridization. Chromosoma 107:272–276
Gawłowska M, Święcicki W, Wolko B (2005) Comparison of genetic maps for two related pea populations (Pisum sativum L.). Pisum Genet 37:19–23
Hole CC, Hardwick RC (1976) Development and control of the number of flowers per node in Pisum sativum L. Ann Bot Lond 40:707–722
Irzykowska L, Wolko B (2004) Interval mapping of QTLs controlling yield-related traits and seed protein content in Pisum sativum. J Appl Genet 45:297–306
Irzykowska L, Wolko B, Święcicki WK (2001) The genetic linkage map of pea (Pisum sativum L.) based on molecular, biochemical and morphological markers. Pisum Genet 33:13–18
Jannink J-L, Jansen R (2001) Mapping epistatic quantitative trait loci with one-dimensional genome searches. Genetics 157:445–454
Li H, Ribaut J-M, Li Z, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Matta NK, Gatehouse JA (1982) Inheritance and mapping of storage protein genes in Pisum sativum L. Heredity 48:383–392
Maurer AR, Ormrod DP, Fletcher HF (1968) Response of peas to environment. IV. Effect of five soil water regimes on growth and development of peas. Can J Plant Sci 48:129–136
Moot DJ, McNeil DL (1995) Yield components, harvest index and plant type in relation to yield differences in field pea genotypes. Euphytica 86:31–40
Ney B, Duthion C, Turc O (1994) Phenological response of pea to water stress during reproductive development. Crop Sci 34:141–146
Poggio SL, Satorre EH, Dethiou S, Gonzalo GM (2005) Pod and seed numbers as a function of photothermal quotient during the seed set period of field pea (Pisum sativum) crops. Eur J Agron 22:55–69
Shia G, Slinkard AE (1977) Relationship of seed shape and cotyledon colour to percent protein of peas. Crop Sci 17:183–184
Święcicki WK (1987) Linkages in Pisum L. IV. The gene DET (determinate growth). Genet Pol 28:75–79
Święcicki W, Święcicki WK (1981) Environmental requirements. In: Legume crops as a source of forage protein (in Polish). Państwowe Wydawnictwo Rolnicze i Leśne, Warszawa
Święcicki WK, Błaszczak P, Hauke J, Mejza S (1981) Inheritance of protein content in pea. III. Correlation between protein content and seed yield. Pisum Newsl 13:52–53
Tar’an B, Warkentin T, Somers DJ, Miranda D, Vandenberg A, Blade S, Bing D (2004) Identification of quantitative trait loci for grain yield, seed protein concentration and maturity in field pea (Pisum sativum L.). Euphytica 136:297–306
Timmerman-Vaughan GM, McCallum JA, Frew TJ, Weeden NF, Russell AC (1996) Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum L.). Theor Appl Genet 93(3):431–439
Timmerman-Vaughan GM, Frew TJ, Butler R, Murray S, Gilipin M, Falloon K, Johnston P, Lakeman MB, Russel A, Khan T (2004) Validation of quantitative trait loci for Ascochyta blight resistance in pea (Pisum sativium L.), using populations from two crosses. Thor Appl Genet 109:1620–1631
Timmerman-Vaughan GM, Mills A, Whitfield C, Frew T, Butler R, Murray S, Lakeman M, McCallum J, Russell A, Wilson D (2005) Linkage mapping of QTL for seed yield, yield components, and developmental traits in pea. Crop Sci 45:1336–1344
Van Ooijen J, Voorrips R (2001) JoinMap version 3.0. Software for the calculation of genetic linkage maps. Plant Research International B.V., Wageningen
VSN International (2010) GenStat for windows, 13th edn. VSN International, Hemel Hempstead. http://www.genstat.co.uk
Wager HG (1974) The effect of subjecting peas to air enriched with carbon dioxide. II. Respiration and the metabolism of the major acids. J Exp Bot 25:338–351
Weeden NF, Święcicki WK, Timmerman-Vaughan GM, Ellis THN, Ambrose M (1996) The current pea linkage map. Pisum Genet 28:1–4
Weeden NF, Ellis TH, Timmerman-Vaughan GM, Święcicki W, Rozov SM, Berdnikov VA (1998) A consensus linkage map for Pisum sativum. Pisum Genet 30:1–4
Windows QTL Cartographer 2.0 (2007) User manual. N.C. State University, Bioinformatics Research Center, Raleigh
Acknowledgments
The work was partially supported by the Polish Ministry of Science and Higher Education grant no. 3 P06A 043 24.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Krajewski, P., Bocianowski, J., Gawłowska, M. et al. QTL for yield components and protein content: a multienvironment study of two pea (Pisum sativum L.) populations. Euphytica 183, 323–336 (2012). https://doi.org/10.1007/s10681-011-0472-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-011-0472-4