Abstract
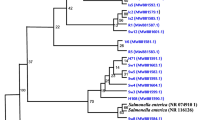
This study investigated antibiotic resistance (ABR) and extended-spectrum ß-lactamases (ESBL) patterns in bacterial isolates collected from the dairy, hotel, meat, and canteen food waste samples. A total of 144 bacterial strains were collected and screened for resistance against 9 standard antibiotics belonging to three generations and ESBL production. The ABR profile of the bacterial isolates was observed against all four major antibiotic groups (aminoglycosides, β-lactams, quinolone, and others), where resistance against cefotaxime (> 70%) and methicillin (> 50%) was high. Though the ABR pattern of strains from dairy waste (> 50%) was high against first-generation antibiotics, the strains from meat waste (> 50%) showed considerable resistance against second- and third-generation antibiotics. ESBL-producing isolates were screened (> 60%, n = 144) through primary identification tests (combined disk test and double disk synergy tests) and further confirmed through Hexa G-minus 23 and 24 and MIC E-stripe following CLSI guidelines. Genes conferring ESBL resistance blaCTX-M, blaSHV, blaOXA, blaTEM, blaKPC genes and multidrug resistance (MDR) mexF gene were detected in the selected isolates with ABR and ESBL traits. Isolates with multidrug ABR and ESBL phenotype were further genotypically identified through 16 s rRNA gene sequencing. The synergy of ABR was detected through the co-expression of ESBL and MDR in isolates with a high occurrence of ABR and ESBL. The results demonstrate the significance of food waste as a natural reservoir of ABR and ESBL-producing pathogens, highlighting the importance of resistance monitoring and its interventions.
Graphical Abstract





Similar content being viewed by others
Data availability
Not applicable.
References
Acolatse, J.E.E., Portal, E.A.R., Boostrom, I., et al. (2022). Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrobial Resistance & Infection Control, 11, 49. https://doi.org/10.1186/s13756-022-01090-2
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research, 48(D1), D517–D525. https://doi.org/10.1093/nar/gkz935
Aslam, B., Khurshid, M., Arshad, M. I., Muzammil, S., Rasool, M., Yasmeen, N., et al. (2021). Antibiotic resistance: One health one world outlook. Frontiers in cellular and infection microbiology, 11, 771510. https://doi.org/10.3389/fcimb.2021.771510
Azab, K. S. M., Abdel-Rahman, M. A., El-Sheikh, H. H., Azab, E., Gobouri, A. A., & Farag, M. M. S. (2021). Distribution of extended-spectrum β-lactamase (ESBL)-encoding genes among multidrug-resistant gram-negative pathogens collected from three different countries. Antibiotics (Basel, Switzerland), 10(3). https://doi.org/10.3390/antibiotics10030247
Bergšpica, I., Kaprou, G., Alexa, E. A., Prieto, M., & Alvarez-Ordóñez, A. (2020). Extended Spectrum β-Lactamase (ESBL) Producing Escherichia coli in Pigs and Pork Meat in the European Union. Antibiotics, 9, 678. https://doi.org/10.3390/antibiotics9100678
Bonnet, R., Sampaio, J. L., Chanal, C., Sirot, D., De Champs, C., Viallard, J. L., Labia, R., & Sirot, J. (2000). A novel class A extended-spectrum beta-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob Agents Chemother, 44(11), 3061–8. https://doi.org/10.1128/AAC.44.11.3061-3068.2000
Bhuwal, A. K., Singh, G., Aggarwal, N. K., Goyal, V., & Yadav, A. (2014). Poly-β-hydroxybutyrate production and management of cardboard industry effluent by new Bacillus sp. NA10. Bioresources and Bioprocessing, 1(1), 9. https://doi.org/10.1186/s40643-014-0009-5
Castanheira, M., Simner, P. J., & Bradford, P. A. (2021). Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrobial Resistance, 3(3), dlab092. https://doi.org/10.1093/jacamr/dlab092
Caudell, M. A., Mair, C., Subbiah, M., Matthews, L., Quinlan, R. J., Quinlan, M. B., et al. (2018). Identification of risk factors associated with carriage of resistant <em>Escherichia coli</em> in three culturally diverse ethnic groups in Tanzania: A biological and socioeconomic analysis. The Lancet Planetary Health, 2(11), e489–e497. https://doi.org/10.1016/S2542-5196(18)30225-0
Chowdhury, S., Ghosh, S., Aleem, M. A., Parveen, S., Islam, M. A., Rashid, M. M., et al. (2021). Antibiotic usage and resistance in food animal production: What have we learned from Bangladesh? Antibiotics (basel, Switzerland), 10(9), 1032. https://doi.org/10.3390/antibiotics10091032
CLSI. (2022). Performance standards for antimicrobial susceptibility testing, 32nd Edition. https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 28 June 2022
Costa, M. M., Cardo, M., Soares, P., Cara d’Anjo, M., & Leite, A. (2022). Multi-drug and β-lactam resistance in Escherichia coli and Food-borne pathogens from animals and food in Portugal, 2014–2019. Antibiotics. https://doi.org/10.3390/antibiotics11010090
Díaz-Gavidia, C., Barría, C., Rivas, L., García, P., Alvarez, F. P., González-Rocha, G., et al. (2021). Isolation of ciprofloxacin and ceftazidime-resistant enterobacterales from vegetables and river water is strongly associated with the season and the sample type. Frontiers in microbiology, 12, 604567. https://doi.org/10.3389/fmicb.2021.604567
Esemu, S. N., Aka, T. K., Kfusi, A. J., Ndip, R. N., & Ndip, L. M. (2022). Multidrug-resistant bacteria and enterobacteriaceae count in abattoir wastes and its receiving waters in Limbe Municipality, Cameroon: Public health implications. BioMed research international. Laboratory for Emerging Infectious Diseases, University of Buea, PO Box 63, Buea, Cameroon. https://doi.org/10.1155/2022/9977371
Foudraine, D. E., Strepis, N., Stingl, C., ten Kate, M. T., Verbon, A., Klaassen, C. H. W., et al. (2021). Exploring antimicrobial resistance to beta-lactams, aminoglycosides and fluoroquinolones in E. coli and K. pneumoniae using proteogenomics. Scientific Reports, 11(1), 12472. https://doi.org/10.1038/s41598-021-91905-w
Gokul, M. S., Dahms, H.-U., Muthukumar, K., Kaviarasan, T., Henciya, S., Vignesh, S., et al. (2019). Chapter 13 - Multimarker pollution studies along the east coast of southern India. In M. Ramkumar, R. A. James, D. Menier, & K. B. T.-C. Z. M. Kumaraswamy (Eds.), (pp. 331–346). Elsevier. https://doi.org/10.1016/B978-0-12-814350-6.00013-6
Gwenzi, W., Chaukura, N., Muisa-Zikali, N., Teta, C., Musvuugwa, T., Rzymski, P., & Abia, A. L. K. (2021). Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics, 10(1). https://doi.org/10.3390/antibiotics10010068
He, Y., Yuan, Q., Mathieu, J., Stadler, L., Senehi, N., Sun, R., & Alvarez, P. J. J. (2020). Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. npj Clean Water, 3(1), 4. https://doi.org/10.1038/s41545-020-0051-0
Jenifer, P., Vijay, R., Rawson, A., Baskaran, N., & Vignesh, S. (2022). Food waste as a reservoir of antibiotic resistant strains: A study on spread of ESBL linked with ABR strains from seafood waste. The Pharma Innovation Journal, 11(8S), 1322–1327. https://www.thepharmajournal.com/special-issue?year=2022&vol=11&issue=8S&ArticleId=14964. Accessed 9 Sept 2022
Jia, J., Zhu, Z., Xue, X., Li, X., & Wang, Z. (2022). Selective pressure governs the composition, antibiotic, and heavy metal resistance profiles of Aeromonas spp. isolated from Ba River in Northwest China. Environmental Science and Pollution Research International. https://doi.org/10.1007/s11356-022-20678-0
Kiehlbauch, J. A., Hannett, G. E., Salfinger, M., Archinal, W., Monserrat, C., & Carlyn, C. (2000). Use of the National Committee for Clinical Laboratory Standards guidelines for disk diffusion susceptibility testing in New York state laboratories. Journal of Clinical Microbiology, 38(9), 3341–3348. https://doi.org/10.1128/JCM.38.9.3341-3348.2000
Kraemer, S. A., Ramachandran, A., & Perron, G. G. (2019). Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms, 7(6), 180. https://doi.org/10.3390/microorganisms7060180
Li, Y., Li, W., Pan, Y., Liu, C., Liang, S., & Zeng, Z. (2022). The emergence and molecular study of methicillin-resistant Staphylococcus aureus ST239, ST59, ST9, and ST630 in food animals, Chongqing, China. Veterinary Microbiology, 265, 109329. https://doi.org/10.1016/j.vetmic.2021.109329
Manaia, C. M., Rocha, J., Scaccia, N., Marano, R., Radu, E., Biancullo, F., et al. (2018). Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environment International, 115, 312–324. https://doi.org/10.1016/j.envint.2018.03.044
Martak, D., Guther, J., Verschuuren, T.D., Valot B, Conzelmann, N., Bunk, S., et al. (2022). Populations of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae are different in human-polluted environment and food items: a multicentre European study. Clinical Microbiology and Infection, 28(3):447.e7-447. https://doi.org/10.1016/j.cmi.2021.07.022
Mir, R., Salari, S., Najimi, M., & Rashki, A. (2022). Determination of frequency, multiple antibiotic resistance index and resistotype of Salmonella spp. in chicken meat collected from southeast of Iran. Veterinary Medicine and Science, 8(1), 229–236. https://doi.org/10.1002/vms3.647
Mummaleti, G., Sarma, C., Kalakandan, S., Sivanandham, V., Rawson, A., & Anandharaj, A. (2021). Optimization and extraction of edible microbial polysaccharide from fresh coconut inflorescence sap: An alternative substrate. Lwt, 138, 110619. https://doi.org/10.1016/j.lwt.2020.110619
Murugan, M. S., Sinha, D. K., Vinodh Kumar, O. R., Yadav, A. K., Pruthvishree, B. S., Vadhana, P., et al. (2019). Epidemiology of carbapenem-resistant Escherichia coli and first report of blaVIM carbapenemases gene in calves from India. Epidemiology and Infection, 147, e159. https://doi.org/10.1017/S0950268819000463
Nagvenkar, G. S., & Ramaiah, N. (2009). Abundance of sewage-pollution indicator and human pathogenic bacteria in a tropical estuarine complex. Environmental Monitoring and Assessment, 155(1–4), 245–256. https://doi.org/10.1007/s10661-008-0432-1
Priyanka Yashwant, C., Krishnmoorthy, S., Loganathan, M. A. R., Anandharaj, A., Baskaran, N., & Vignesh, S. (2021). Biotransformation of food waste to starter culture biomass: An investigation of antibiotic resistance-free lactic acid bacteria from dairy and household food waste. The Pharma Innovation Journal, 10(10S), 601–607. https://www.thepharmajournal.com/special-issue?year=2021&vol=10&issue=10S&ArticleId=8203. Accessed 11 Aug 2022
Qiao, M., Ying, G.-G., Singer, A. C., & Zhu, Y.-G. (2018). Review of antibiotic resistance in China and its environment. Environment International, 110, 160–172. https://doi.org/10.1016/j.envint.2017.10.016
Rajivgandhi, G., Maruthupandy, M., Ramachandran, G., Priyanga, M., & Manoharan, N. (2018). Detection of ESBL genes from ciprofloxacin resistant Gram negative bacteria isolated from urinary tract infections (UTIs). Frontiers in Laboratory Medicine, 2(1), 5–13. https://doi.org/10.1016/j.flm.2018.01.001
Ramachandran, G., Rajivgandhi, G. N., Chackaravarthi, G., Kanisha, C. C., Siddiqi, M. Z., Alharbi, N. S., et al. (2021). Isolation and molecular identification of extended spectrum beta-lactamase producing bacteria from urinary tract infection. Journal of Infection and Public Health, 14(12), 1911–1916. https://doi.org/10.1016/j.jiph.2021.10.016
Sanganyado, E., & Gwenzi, W. (2019). Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Science of the Total Environment, 669, 785–797. https://doi.org/10.1016/j.scitotenv.2019.03.162
Sinha, R. (2018). Dairy farm wastewater increases presence of antibiotic resistant genes in soil. DowntoEarth. https://www.downtoearth.org.in/news/health/irrigation-with-farm-wastewater-enhances-presence-of-antibiotic-resistance-genes-in-soil-60451. Accessed 10 Apr 2022
Sivakumar, M., Abass, G., Vivekanandhan, R., Singh, D. K., Bhilegaonkar, K., et al. (2021). Extended-spectrum beta-lactamase (ESBL) producing and multidrug-resistant Escherichia coli in street foods: A public health concern. Journal of Food Science and Technology, 58(4), 1247–1261. https://doi.org/10.1007/s13197-020-04634-9
Song, H.-J., Kim, S.-J., Moon, D. C., Mechesso, A. F., Choi, J.-H., Kang, H. Y., et al. (2022). Antimicrobial resistance in Escherichia coli isolates from healthy food animals in South Korea, 2010–2020. Microorganisms. https://doi.org/10.3390/microorganisms10030524
Song, J., Oh, S.-S., Kim, J., Park, S., & Shin, J. (2020). Clinically relevant extended-spectrum β-lactamase–producing escherichia coli isolates from food animals in South Korea. Frontiers in Microbiology, 11. https://doi.org/10.3389/fmicb.2020.00604
Srinivasan, K., & Buys, E. M. (2019). Insights into the role of bacteria in vitamin A biosynthesis: Future research opportunities. Critical Reviews in Food Science and Nutrition. https://doi.org/10.1080/10408398.2018.1546670
Tomaszewska, M., Bilska, B., Tul-Krzyszczuk, A., & Kołożyn-Krajewska, D. (2021). Estimation of the scale of food waste in hotel food services—A case study. Sustainability, 13(1). https://doi.org/10.3390/su13010421
Vignesh, S., Muthukumar, K., & James, R. A. (2012). Antibiotic resistant pathogens versus human impacts: A study from three eco-regions of the Chennai coast, southern India. Marine Pollution Bulletin, 64(4), 790–800. https://doi.org/10.1016/j.marpolbul.2012.01.015
Vignesh, S., Dahms, H.-U., Emmanuel, K. V., Gokul, M. S., Muthukumar, K., Kim, B.-R., & James, R. A. (2014). Physicochemical parameters aid microbial community? A case study from marine recreational beaches, Southern India. Environmental Monitoring and Assessment, 186(3), 1875–1887. https://doi.org/10.1007/s10661-013-3501-z
Vignesh, S., Dahms, H. U., Kumarasamy, P., Rajendran, A., Kim, B. R., & James, R. A. (2015). Microbial effects on geochemical parameters in a tropical river basin. Environmental Processes, 2(1), 125–144. https://doi.org/10.1007/s40710-015-0058-6
Vignesh, S., Dahms, H. U., Muthukumar, K., Vignesh, G., & James, R. A. (2016). Biomonitoring along the tropical Southern Indian Coast with multiple biomarkers. PLOS One, 11(12), e0154105. https://doi.org/10.1371/journal.pone.0154105
Vignesh, S, Muthukumar, K., Gokul, M. S., & James, R. A. (2013). Microbial pollution indicators in the Cauvery River, Southern India. In M. Ramkumar (Ed.), On a Sustainable Future of the Earth’s Natural Resources (pp. 363–376). Berlin, Heidelberg: Springer Berlin Heidelberg. https://doi.org/10.1007/978-3-642-32917-3_20
Vijay, R., Srinivasan, K., Arunkumar, A., Baskaran, N., Ashish, R., & Vignesh, S. (2021). Enhanced exopolysaccharide production from food waste as a substrate through fed-batch FMN: An exploratory investigation of fluoride resistant bacteria. The Pharma Innovation, SP-10(10), 594–600.
Wang, M., Yao, M., & Zhu, Y. (2022). Antibiotic resistance genes and antibiotic sensitivity in bacterial aerosols and their comparisons with known respiratory pathogens. Journal of Aerosol Science, 161, 105931. https://doi.org/10.1016/j.jaerosci.2021.105931
Yashwant, C. P., Rajendran, V., Krishnamoorthy, S., Nagarathinam, B., Rawson, A., Anandharaj, A., & Sivanandham, V. (2022). Antibiotic resistance profiling and valorization of food waste streams to starter culture biomass and exopolysaccharides through fed-batch fermentations. Food Science and Biotechnology. https://doi.org/10.1007/s10068-022-01222-9
Zalewska, M., Błażejewska, A., Czapko, A., & Popowska, M. (2021). Antibiotics and antibiotic resistance genes in animal manure – Consequences of its application in agriculture. Frontiers in Microbiology, 12. https://doi.org/10.3389/fmicb.2021.610656
Acknowledgements
The authors express their sincere gratitude to the National Institute of Food Technology, Entrepreneurship and Management (NIFTEM)—Thanjavur, Thanjavur, Tamil Nadu, for providing the facilities and the Department of Science & Technology (DST-SERB-SRG/2021/001005) for funding support to carry out the study.
Funding
Dr. Vignesh Sivanandham has received a research grant from the Department of Science & Technology, Science & Engineering Research Board (DST-SERB-SRG/2021/001005).
Author information
Authors and Affiliations
Contributions
Jenifer performed all the experiments and generated data; Srinivasan wrote the manuscript and analyzed the data; Baskaran performed the molecular analysis and reviewed the manuscript. Vignesh conceptualized the study, designed the experiment, and supervised and reviewed the research work.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to publish
The authors give consent for the publication of the manuscript, which includes images and tables within the text to be published in the Journal and Article.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
As the Corresponding author, I confirm that the manuscript has been read and approved for submission by all the named authors. We declare that this manuscript is original, has not been published before, and is not currently being considered for publication elsewhere.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Periasamy, J., Krishnamoorthy, S., Nagarethinam, B. et al. Food wastes as a potential hotspot of antibiotic resistance: synergistic expression of multidrug resistance and ESBL genes confer antibiotic resistance to microbial communities. Environ Monit Assess 195, 783 (2023). https://doi.org/10.1007/s10661-023-11335-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11335-1