Abstract
Banana is a staple food and cash crop grown in East and Central Africa (ECA). The main banana varieties grown in ECA are the East African highland bananas (EAHB), although dessert/beer bananas such as Sukari Ndizi, Kayinja (Pisang Awak) and Gros Michel are also produced due to their high value at local markets. The Fusarium wilt fungus Fusarium oxysporum f. sp. cubense (Foc) causes disease of susceptible dessert/beer bananas, which significantly reduces yields. Banana Fusarium wilt is managed by excluding the pathogen from disease-free areas and by planting disease-resistant varieties in infested fields. Six phylogenetically closely-related vegetative compatibility groups (VCGs) of Foc, VCGs 0124, 0125, 0128, 01212, 01220 and 01222 are present in ECA, which all group together in Foc Lineage VI. Rapid and accurate detection of Foc Lineage VI strains is thus important to prevent its spread to disease-free areas. In this study, molecular markers specific to Foc Lineage VI were therefore developed. Primer sets were then combined in a multiplex PCR assay, and validated on a worldwide population of 623 known Foc isolates, other formae speciales and non-pathogenic Fusarium oxysporum isolates. The Foc Lineage VI multiplex PCR was used to identify Foc isolates collected in banana fields at five locations in Uganda and Tanzania. Foc Lineage VI DNA was detected at a concentration as low as 0.1 ng/μl, both in the absence and presence of banana DNA, and can therefore be used as an accurate diagnostic tool for Foc Lineage VI strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana (Musa spp.) is produced as a staple food and cash crop in Uganda, Kenya, Tanzania, Burundi, Rwanda and the Democratic Republic of the Congo (DRC). This area is collectively known as East and Central Africa (ECA), and produced 16.8 million of the 153.2 million tons of bananas produced worldwide in 2017 (FAOSTAT 2017). Bananas in ECA are mainly grown for local consumption and trade, and are valued at an estimated US$ 4.80 billion (FAOSTAT 2016) annually. In Uganda, Burundi and Rwanda, annual consumption levels per capita are approximately 131, 233 and 297 kg, respectively (FAOSTAT 2013). This is substantially more than the average annual global consumption of 20 kg per person (FAOSTAT 2013). Yet, the yields in ECA countries are much lower, often less than 10 tons per ha year−1, compared to 50 tons per ha year−1 achieved by leading banana-producing countries such Costa Rica and Indonesia (FAOSTAT 2017). The low yields in ECA are primarily caused by poor soil fertility, water shortage, pests and diseases, poor management and socio-economic constraints (Karamura et al. 1998; Wairegi et al. 2010; Van Asten et al. 2011).
Fusarium wilt is a soil-borne disease caused by the fungus Fusarium oxysporum Schlect. f. sp. cubense (E.F. Smith) Snyder and Hansen (Foc) (Stover 1962). The disease was first reported in ECA on the ABB banana varieties Bluggoe and Pisang Awak in Tanzania, Uganda and Kenya in the 1950s and 1960s, and in Rwanda and Burundi in the 1980s (Gatsinzi and Sebasigari 1989; Ploetz et al. 1990). Foc strains causing disease to bananas in ECA belong to Foc races 1 and 2, and include vegetative compatibility groups (VCGs) 0124, 0125, 0128, 01212, 01220 and 01222. VCG 0120, 01213/16 and 01214 are also present on the African continent, but occur only in South Africa, northern Mozambique and Malawi, respectively (Blomme et al. 2013; Butler 2013; Karangwa et al. 2017). The six VCGs reported in ECA are phylogenetically related and all cluster together in Foc Lineage VI (Fourie et al. 2009). VCGs within Lineage VI often form complexes, of which the VCG 0124–01222 complex is the most common in the region (Karangwa et al. 2017).
Banana production in ECA is dominated by East African Highland bananas (EAHB), which are unique to the region (Karamura et al. 1998; Gold et al. 2002). Triploid EAHB bananas (AAA), commonly known as Matoke bananas, are resistant to Foc Lineage VI strains (Kangire et al. 2000; Tushemereirwe et al. 2000). However, diploid bananas (AA) grown in some parts of Kenya (Onyango et al. 2011), and known as Muraru bananas, are susceptible to Fusarium wilt (Kung’u and Jeffries 2001). The susceptibility of Mchare (AA) bananas grown in the Kilimanjaro region in Tanzania (De Langhe et al. 2001) has not previously been reported. Foc Lineage VI strains also affect a number of exotic cultivars introduced onto the continent in the twentieth century, including dessert bananas such Gros Michel (AAA), Sukari Ndizi (Apple or Silk-like AAB banana) and the beer banana Kayinja (Pisang Awak, ABB) (Karamura et al. 1998; Ploetz and Pegg 2000). Sukari Ndizi (also known as Kamaramasenge) is becoming increasingly popular for wine and beer production in Rwanda. Farmers often grow dessert/beer cultivars in mixed systems with EAHB. Due to the cost of tissue culture bananas, suckers are used to establish new plantations (Karamura et al. 1996; Karamura et al. 1998; Karangwa et al. 2016). As a consequence, Fusarium wilt is continuously spreading (Blomme et al. 2013; Karangwa et al. 2016) and is considered as an important disease in the region (Tushemereirwe et al. 2004; Nkuba et al. 2015).
Banana Fusarium wilt can be prevented by planting disease-free bananas in Foc-free fields, and by growing resistant varieties in Foc-infested fields. For instance, Cavendish bananas were used to replace the susceptible Gros Michel bananas in Latin America and Africa when it became difficult to continue growing Gros Michel for the international banana export industry (Stover 1962). Other disease management strategies, however, have had a limited impact on disease reduction (Ploetz 2015a). Prevention of spread also involves effective phytosanitary measures and the early detection of the pathogen in contaminated plants, water and soil (Miller et al. 2009; Carter et al. 2010). The development of resistant banana varieties is a slow and expensive process and requires adequate screening in fields contaminated with all variants of the fungus present in a geographical area. For instance, the supposedly resistant banana hybrid “Golden Beauty” succumbed to Fusarium wilt when it was released in Honduras by the Imperial College of Tropical Agriculture (ICTA) in 1928 (Ploetz and Churchill 2011). This failure was attributed to the lack of consideration of Foc variation during the screening process (Stover and Buddenhagen 1986). An accurate diagnostic tool for Foc variants is, therefore, needed for the management of banana Fusarium wilt.
Race classification has been used to distinguish among Foc strains, and is based on the pathogen’s ability to cause disease to a differential set of banana cultivars. There are three Foc races; race 1, race 2 and race 4, causing disease to Gros Michel, Bluggoe and Cavendish cultivars, respectively (Stover 1962; Kung’u and Jeffries 2001; Ploetz 2006, 2015a). The identification of Foc races is, however, not always reliable as some banana varieties are predisposed to infection by Foc in unfavourable environments (Ploetz 2015a). For instance, some Foc strains isolated from Gros Michel (differential cultivar of Foc race 1) cause disease to Bluggoe (differential cultivar to Foc race 2) and vice versa (Kung’u and Jeffries 2001). VCGs are now used to identify Foc strains, but VCG testing is a time-consuming technique and and gives no indication of genetic relatedness/distance between different VCGs. Molecular diagnostics can speed up identification of Foc strains and are also more accurate and reproducible than the VCG technique and pathogenicity tests.
Molecular markers have been developed for the identification of Foc race 4 strains (Lin et al. 2009; Dita et al. 2010; Lin et al. 2013; Zhang et al. 2013; Aguayo et al. 2017), but are not specific for Foc Lineage VI strains. The objective of this study was to develop a PCR-based diagnostic tool targeting Foc Lineage VI strains. The availability of molecular markers for Foc Lineage VI could support quarantine regulations in ECA and assist in the screening of banana varieties for resistance to Foc in the region. They can also be of value in other geographic areas where Foc Lineage VI strains affect bananas.
Materials and methods
Fungal isolates used and DNA extraction
Eighty-four isolates of F. oxysporum, including Foc isolates representing the 24 known VCGs, other formae speciales and non-pathogenic isolates, were used to develop a Lineage VI-specific primer set (Table 1). Additionally, 623 fungal isolates from a worldwide Foc collection were used to validate the markers (Online Resource 1). The isolates are all single-spored cultures and are maintained in the culture collection of Stellenbosch University’s Department of Plant Pathology (USPP) in 15% glycerol at −80 °C.
DNA was extracted after growing the cultures on potato dextrose agar (PDA) in 90-mm-diameter Petri dishes at 25 °C for 7 days. Mycelia were scraped off the media with sterile scalpel blades, and transferred into 2 ml Eppendorf tubes. DNA was extracted using the Wizard SV Genomic DNA Purification Systems kit according to the manufacturer’s instructions (Promega, Madison, USA) and stored at −20 °C until use.
Primer design
Multiple sequence alignment databases of translation elongation factor-1 alpha (TEF-1α) (Fourie et al. 2009) and DNA-directed RNA polymerase III subunit (RPC2) gene regions (unpublished data) available at Stellenbosch University were used to identify single nucleotide polymorphisms specific to Foc Lineage VI.
DNA sequence
DNA of each fungal isolate was first sheared into 300-bp fragments using a Covaris S2 (Woburn, MA, USA) instrument, and the DNA fragments used to prepare libraries for sequencing according to instructions provided in the NEBNext DNA library preparation kit for Illumina (New England Biolabs, Ipswich, MA, USA). The quality and quantity of the DNA fragments were verified using an Agilent Bio-analyser (Santa Clara, CA, USA). The DNA of the 84 isolates was sequenced in two lanes of an Illumina Hiseq 2000 (San Diego, CA, USA), and raw reads were pre-processed with Trimmomatic (Bolger et al. 2014). Trimmed reads were then mapped to 10 selected conserved single copy orthologous genes provided by Li-Jun Ma (University of Massachusetts, MA, USA). Mapping was done in CLC genomics workbench version 11 (Qiagen, Hilden, Germany) using default parameters to create a consensus sequence of each gene for each isolate.
Sequence alignment and primer design.
Multiple sequence alignments of both gene areas were performed using MAFFT software version 5.85 (Katoh et al. 2005), and visualized with MEGA software for easy identification of single nucleotide polymorphisms (SNPs) specific to Foc Lineage VI. A primer set was designed based on unique SNPs in RPC2 gene region that are specific to Foc Lineage VI isolates. Primers were designed with Primer3 software using the following oligonucleotide properties: Melting temperatures between 55 and 65 °C, primer length between 18 and 25 bp, GC content between 40 and 60%, and 3′-end sequence and self-dimers checking(Untergrasser et al. 2012).
Specificity testing of the primer set
Each primer set was first tested on seven isolates representing seven Foc lineages (Fourie et al. 2009). When the primer set proved to be specific, the testing was extended to a collection of 84 F. oxysporum isolates, including representatives of the 24 Foc VCGs, other formae speciales and non-pathogenic isolates (Table 1).
Each PCR reaction consisted of 25 μl including 12.5 μl of Kapa ready mix (1 U per 50-μl reaction of Kapa Taq DNA Polymerase, 0.2 mM of each dNTP, 1.5 mM MgCl2 and stabilisers) (Kapa biosystems, Cape Town, South Africa), 9.5 μl nuclease-free water, 0.5 μl of each of the 10 μM primers, and 2 μl of the 10 ng/μl DNA template. PCR conditions consisted of an initial denaturation step at 95 °C for 2 min, followed by 30 cycles for denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s and elongation at 72 °C for 30 s. The final extension step was set at 72 °C for 5 min.
Multiplex assay, validation and sensitivity testing
The primer set developed in the current study was combined in a multiplex PCR assay with the primer set FocLin6bF (5’-CGACAATGAGCTTATCTGCCATT-3′) and FocLin6bR (5’-CATCGAGGTTGTGAGAATGGA-3′) (Karangwa 2015). FocLin6bF/R, also specific to Foc Lineage VI, was designed from the translation elongation factor-1 alpha (TEF-1α) gene region and amplified a DNA fragment of 300 base pairs (bp). In the multiplex PCR reaction, 12 μl of Kapa ready mix, 9.0 μl nuclease-free water, 0.5 μl of each of the 10 μM primers and 2 μl of 10 ng/μl DNA template were used in the 25 μl reaction mix. PCR conditions were set as described above. The specificity of the multiplex assay was then validated against a worldwide population of 623 isolates of F. oxysporum isolates including Foc isolates, other formae speciales and non-pathogenic isolates from different regions of the world (Online Resource 1). The sensitivity of the primer sets in the multiplex PCR was also tested on decreasing concentrations from 5 to 0.01 ng/μl of fungal DNA and in the presence of 50 ng/μl banana DNA. Positive controls were included for the detection of false negatives.
Identifying Foc isolates in five locations of ECA
Collection of samples
Eighty-nine banana farms were surveyed in five locations in two ECA countries in 2016 and 2017, and 220 samples collected from banana plants showing typical symptoms of Fusarium wilt. The locations were considered major banana-growing regions in the two countries, and included Kawanda and Mbarara in Uganda, and Arusha, Mbeya and Bukoba in Tanzania (Online Resource 2). Banana plants expressing typical external symptoms such as leaf yellowing and collapsing were selected, and their pseudostems split open to expose the internal symptoms of Fusarium wilt. Three to five vascular strands per plant were sampled from diseased pseudostems, placed in a paper towel and kept in paper envelope bags. Samples were transported and stored at room temperature prior to isolation. Information on cultivation history, disease incidence, affected varieties and GPS coordinates were also recorded. The disease incidence was evaluated using two approaches because susceptible cultivars to Fusarium wilt were grown in mixtures with resistant cultivars including EAHB, plantains and Cavendish types. For the first approach the disease incidence was determined as a percentage of diseased plants out of the total number of susceptible plants. In the second approach, the percentage of diseased plants out of 30 plants randomly selected at the farm, including all banana types, was used.
Fungal isolation, morphological identification and storage
Fungal isolation was carried out at USPP. Two to three fragments of about 5 mm each per sample were excised from the vascular strands and plated onto PDA amended with streptomycin (4 mg/L). Cultures were incubated at 25 °C for 5–7 days, purified and single-spored, and maintained in 15% glycerol at −80 °C in the CAV culture collection of USPP. All isolates were grown on PDA and carnation leaf agar (CLA) for cultural and morphological identification according to Leslie and Summerell (2006). Isolates considered as F. oxysporum species were further tested with the multiplex PCR developed for Foc Lineage VI. Non-Fusarium species, which were culturally and morphologically characterised, were not further investigated.
Molecular identification of Fusarium species
All F. oxysporum isolates collected from diseased bananas in ECA were first identified using the multiplex PCR, both as single-spore cultures and in infected strands collected from diseased banana plants. Genomic DNA from banana tissue was extracted using a NucleoSpin® Plant II kit (Macherey-Nagel, Düren, Germany). The PCR reaction for in planta detection was carried out as follows: A total reaction volume of 40 μl was used that contained 4 μl 10x NH4 reaction buffer, 2 μl of 50 mM MgCl2, 0.8 μl of 250 μM dNTPs, 2 μl of 20 ng/ml bovine serum albumin (BSA), 0.13 μl of 5 u/μl Taq polymerase, 0.8 μl of each of the10 ng/μl primers, 25.87 μl nuclease-free water and 2 μl of DNA template. PCR conditions were similar to those described for the amplification of fungal DNA.
Vegetative compatibility characterization of Foc
The identity of all Foc cultures recovered from diseased banana pseudostems was confirmed by VCG analysis (Leslie and Summerell 2006). The isolates were first cultured on PDA at 25 °C for 7 days, and small plugs of the developing mycelia transferred to minimal media amended with 1–3% KClO3. The isolates were then incubated at 25 °C for 7–21 days, and ClO3-resistant mutants that developed were transferred to minimal media slants. Colonies with sparse mycelial growth were considered nitrate non-utilizing (nit)-mutants, and phenotyped as nit-1, nit-3 or Nit-M mutants (Leslie and Summerell 2006). Nit-1 mutants of unknown isolates were then paired with the Nit-M mutant of the same isolate on minimal medium to confirm heterokaryon self-compatibility, and with the Nit-M of the tester isolate to determine their VCG identity. Heterokaryons are identified when complementary hyphae of nit-mutants anastomose to form a fluffy wild-type (Leslie and Summerell 2006). If fluffy wild-type growth occur between the mutants, the unknown isolate will belong to the same VCG as the tester strain. Heterokaryon self-incompatible strains are identified when the nit-1 mutants do not pair with their own Nit-M mutants.
Results
Primers and their specificity to the lineage VI
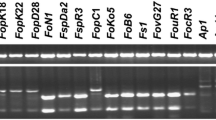
The two best performing primer sets were FocLin6bF (5’-CGACAATGAGCTTATCTGCCATT-3′) and FocLin6bR (5’-CATCGAGGTTGTGAGAATGGA-3′) that amplified a DNA fragment of 300 bp within the TEF-1α gene region, and FocLinVI-F (5’-AGGGACTGGATTTCTACCCT-3′) and FocLinVI-R (5’-GTGTCACTTGGTCCTCGTAT-3′) that amplified a 1002 bp DNA fragment in the RPC2 gene region. For the former, the forward and reverse primers were designed to include SNPs specific to Foc Lineage VI at positions 60 and 356 bp, respectively (Fig. 1). For the latter, the forward primer included a SNP exclusive to Foc Lineage VI at the 3′-end at position 1007, while the reverse primer has two specific SNPs at positions 1989 and 1992 at the 3′-end (Fig. 2). When the specificity of both primer sets was tested on seven Foc strains it exclusively amplified CAV 2260, an isolate that representing VCG 0124 in Foc Lineage VI, but not the isolates that represent the other six Foc lineages (Fig. 3). When the specificity test was extended to 84 isolates (Table 1), both primer sets consistently amplified only isolates associated with Foc Lineage VI.
Sequence alignment of the DNA-directed RNA polymerase III subunit region (RPC2) of Fusarium oxysporum f. sp. cubense (Foc) isolates representing 24 vegetative compatibility groups. The dark blocks show single nucleotide polymorphisms exclusive to Foc Lineage VI. The forward primer was designed to include the Lineage VI-specific SNP at position 1007, while the reverse primer was designed to include the Lineage VI specific SNP at position 1989 and 1992
Specificity testing of the two primer sets individually and combined in a multiplex PCR assay for Fusarium oxysporum f. sp. cubense (Foc) Lineage VI. Left: A 300 bp fragment amplified by the FocLin6b primer set, Middle: A 1002 bp fragment amplified by the FocLin VI primer set, and Right: Both 300 bp and 1002 bp fragments of Foc amplified by the two primer sets in a multiplex PCR assay. Lanes 1–8: Isolates CAV 980, 618, 789, 871, 968, 2260, 317 and a non-template control; representing Foc Lineage IV, III, V, VIII, VI and F. oxysporum f. sp. melonis, respectively.
Multiplex assay, validation and sensitivity testing
The two primer sets, FocLinVI-F/R and FocLin6bF/R, successfully amplified the two expected DNA fragments of Foc Lineage VI (CAV 2260) in a multiplex assay (Fig. 3). When tested against a worldwide population of 623 fungal isolates the two primer sets only amplified Foc Lineage VI isolates. There was no amplification of other Foc lineages, formae speciales or non-pathogenic F. oxysporum isolates. The primers could detect fungal DNA as low as 0.1 ng/μl in the absence and presence of 50 ng of banana DNA, and were sensitive enough to detect the isolates of the Foc Lineage VI in diseased plants (Fig. 4).
Sensitivity testing of the FocLin6F/R and FocLinVI-F/R markers for the detection of Fusarium oxysporum f. sp. cubense Lineage VI. Left: amplification of Foc DNA at decreasing concentrations, with lanes 1 to 5 corresponding to 5; 2; 1; 0.1 and 0.01 ng/μl, respectively. Middle: Amplification of Foc DNA at decreasing concentrations in the presence of banana DNA, with lanes 1 to 5 corresponding to 5; 2; 1; 0.1 and 0.01 ng/μl mixed each with 50 ng of banana DNA. Right: Detection of Foc in infected planting materials. Lanes 1 to 7 represent seven samples collected from seven ‘Gros Michel’ plants artificially infected with Foc Lineage VI isolates CAV 2576, 2595, 2599, 2640, 2750, 2858 and 2880
Banana varieties affected by Foc and Fusarium wilt incidence
Fusarium wilt was found in susceptible banana cultivars such as Sukari Ndizi (AAB), Pisang Awak (ABB) and Mchare (AA) bananas at the five locations surveyed in Uganda and Tanzania, but not on plantain and EAHB bananas. Foc was mostly recovered from Sukari Ndizi in Uganda and Sukari Ndizi, Pisang Awak and Mchare in Tanzania. The highest incidence of Fusarium wilt was recorded in Mbeya, Tanzania. Sixty-four percent of plants from susceptible varieties were affected by the disease, but when all the banana varieties were taken into account, the disease incidence dropped to 17%. At Kilimanjaro in Tanzania, 29% of Mchare bananas were affected by Fusarium wilt, and in Uganda (Mbarara and Kawanda) the incidence was highest in farms planted with Sukari Ndizi. In the two countries 49, 29 and 13% of the Foc isolates were collected from Sukari Ndizi, Pisang Awak and Mchare bananas, respectively (Table 2). The remaining 9% of isolates were collected from Khom, Safeti Velchi, Embu, Figue Pomme Geante, Kisubi, Kataraza, Kikonjwa, Gros Michel, Home, Igyinga and Kijoge bananas (Table 2). Fusarium wilt was not observed on Matoke and Cavendish bananas.
Identifying F. oxysporum isolates from ECA
Two hundred and twenty isolates collected in Tanzania and Uganda were identified as F. oxysporum using cultural and morphological characteristics. These isolates showed typical cottony growth with a white to violet colour on PDA. The F. oxysporum isolates produced elliptical-shaped microconidia, slightly-curved macroconidia and round chlamydospores produced alone or in pairs on CLA. The isolates all tested positive for Foc Lineage VI multiplex PCR. VCG analysis further characterised these isolates as as members of VCG 0124 (22 isolates), VCG 0125 (5), VCG 0128 (5), VCG 01212 (64), VCG 01220 (1), VCG 01222 (18) and VCG complexes thereof (100) (Table 2). Five isolates were heterokaryon self-incompatible (Online Resource 2).
VCG 0124 and the VCG complexes 0124/22 and 0124/8/22 were dominant in Mbarara, Kawanda, Kagera and Arusha (Table 2, Fig. 5). VCG 01212 was the most abundant VCG found in Mbeya, Tanzania, but it was not found in Uganda (Table 2, Fig. 5). VCG 01220 was the least represented (Online Resource 2) with a single isolate from Kagera (Pisang Awak). All the Foc Lineage VI VCGs were recovered from Pisang Awak, while Sukari Ndizi hosted all the Foc Lineage VI VCGs except VCG 01220. The VCGs recovered from Mchare were VCGs 0124, 01212, and VCG complexes 0124/22 and 0124/8/22 (Table 2, Online Resource 2).
Discussion
VCGs in Foc Lineage VI are the only strains of the Fusarium wilt fungus found in ECA (Kung’u and Jeffries 2001; Blomme et al. 2013; Karangwa et al. 2017). They also occur commonly in many other banana-growing countries, particularly in India and Asia (Mostert et al. 2017). Foc Lineage VI isolates are characterized by pathogenicity testing and VCG analysis, and cannot be distinguished from other Foc or non-pathogenic F. oxysporum strains by morphological differences (Ploetz 2000). Pathogenicity testing and VCG analysis, however, are laborious and time consuming. Disease development is also influenced by the environment, while VCG analysis requires the availability of reference strains, which presents a quarantine risk. Vegetative self-incompatibility can occur when a particular strain’s nit-mutants do not pair with itself or with Foc VCG testers, making VCG characterisation unreliable (Bentley et al. 1998; Lodwig et al. 1999; Mostert et al. 2017; Karangwa et al. 2017). The markers developed in this study, therefore, present a valuable tool that can be used to speed up the identification of Foc Lineage VI strains and to guide the management of banana Fusarium wilt in ECA. It can also be used to accurately identify Foc Lineage VI strains in infected planting material.
This study confirms for the first time that Foc Lineage VI strains can affect Mchare bananas in Tanzania. In addition, popular dessert/beer banana varieties grown by smallholder farmers for local markets in ECA, such as Sukari Ndizi, Kayinja and Gros Michel (Kung’u and Jeffries 2001; Blomme et al. 2013; Karangwa et al. 2017) were also affected, but not the internationally traded Cavendish variety. Within ECA, Cavendish bananas are regarded as inferior to ‘Pisang Awak’, which are highly valued for the production of wine, beer and liquors due to their higher sugar content, as well as Sukari Ndizi and Gros Michel that are highly prized as a table banana in urban areas (Karangwa et al. 2016). Some ECA countries have even envisaged the export of Sukari Ndizi, but were discouraged by the damage caused by Fusarium wilt on the cultivar in the region (Van Asten et al. 2010; Karangwa et al. 2016). Considering the current lack of resistant genotypes to replace Mchare cooking bananas and popular dessert/beer/wine bananas in the region, the production of such bananas relies on planting of disease-free plants in pathogen-free areas, as well as pathogen exclusion. These include the use of plants produced in tissue culture and the enforcement of quarantine regulations. The spread of Foc to new areas can be prevented by restricting the movement of diseased vegetative material, and by the early identification and rogueing of infected plants. In this regard, the molecular markers developed in this study could be a valuable tool to accurately and rapidly detect Foc Lineage VI isolates in diseased planting materials.
Foc TR4 is considered the most destructive Foc race known (Ploetz 2015b). It was first detected in Indonesia and Malaysia in 1990, and has since spread throughout Asia and to the Middle East (Viljoen et al. 2019). Foc TR4 was detected in northern Mozambique in 2013 (Beed 2013)and there are significant concerns that the fungus may spread to neighbouring countries such as Tanzania, Malawi, Zimbabwe and Zambia through the transboundary movement of planting material. Commercial Cavendish plantations are now expanding in east and southern African countries for regional markets and export to the Middle East. In addition to Cavendish bananas, Foc TR4 can threaten Foc Lineage VI-susceptible varieties in Africa such as Mchare, Pisang Awak and Sukari Ndizi. Symptoms caused by Foc TR4 do not differ visually from those caused by Foc races 1 or 2. Therefore, molecular markers of Foc Lineage VI developed in this study, and available Foc TR4 markers (Lin et al. 2009; Dita et al. 2010; Lin et al. 2013; Zhang et al. 2013; Aguayo et al. 2017), should be combined and made available to extension services and agricultural research centres to enhance their capacity to diagnose Fusarium wilt pathogens in the East and Central African region.
Sequence variability among closely-related fungal species is essential for marker development. The alignment of the TEF-1α and RPC2 gene sequences of Foc demonstrated that Foc Lineage VI isolates contain SNPs useful for marker design. The TEF-1α region is a conserved and highly informative gene region in Fusarium species, and is extensively used for phylogenetic and evolutionary studies, as well as for molecular identification (Geiser et al. 2004; O’Donnell et al. 2010). TEF-1α gene sequences have been used to differentiate phylogenetic lineages of Foc (O'Donnell et al. 1998; Fourie et al. 2009). RPC2 has also proved to be phylogenetically informative for the F. oxysporum species complex (Epstein et al. 2017), and was a good candidate gene region for marker development in this study. The use of two primer sets in a multiplex PCR can prevent the undesirable event of obtaining false negatives, as each amplicon serves as a control for the other fragment (Edwards and Gibbs 1994; Markoulatos et al. 2002). Degraded DNA can be among the common causes of failed PCR amplification that results in false negatives (Schrader et al. 2012).
The Foc Lineage VI-specific markers in this study could potentially also be altered to detect the fungus using quantitative real-time PCR (qPCR). The in planta sensitivity test performed in this study showed that Foc Lineage VI markers could detect as little as 0.1 ng/μl of the fungus in the presence of banana DNA and in planting material. qPCR markers may thus be optimised for the detection of Foc Lineage VI in plants, water and soil. It could also be used to screen banana varieties for resistance to Fusarium wilt if pathogen DNA in infected tissues (roots and rhizomes), detected by qPCR, correlates to resistance in banana to Foc. Jiménez-Fernández et al. (2011) and Gramaje et al. (2013), for instance, have used in planta quantification of F. oxysporum f. sp. ciceris causing Fusarium wilt in chickpea and Verticillium dahliae Klebahn causing Verticillium wilt in olive, respectively, to evaluate genotype resistance. They found that the amount of pathogen DNA quantified at the same time point in each genotype correlated well with the resistance of genotypes.
The incidence of Fusarium wilt on banana farms in ECA depended on whether susceptible or resistant cultivars were grown. The incidence was high in farms dominated by Sukari Ndizi, Pisang Awak and Mchare bananas, but was low when susceptible cultivars were mixed with Matoke (EAHB-AAA) bananas. The low incidence found in mixed cultivar settings is consistent with previous reports (Karangwa et al. 2016), although the causes for disease reduction have not been investigated yet. The incidence of 29% in Mchare (EAHB-AA) bananas grown in monoculture at Arusha and Kilimanjaro poses a significant threat to the livelihoods of people in the region. Mchare are cooking bananas preferred by growers in the region (De Langhe et al. 2001). Mchare bananas are genetically similar to edible diploid bananas (AA) grown in Kenya with the generic name ‘Muraru’ (Perrier et al. 2018). Muraru bananas were previously reported to be susceptible to Foc VCGs 0124 and 01220 (Kung’u and Jeffries 2001).
Foc VCG 01212 dominated the Foc population in Mbeya and has been reported to occur in ECA only (Karangwa et al. 2017), indicating the potential evolution of Foc Lineage VI in the region. When compared to other VCGs in Lineage VI, VCG 01212 has little to no sequence variation in conserved sequences like TEF, the intergenic spacer (IGS), RNA polymerase II largest subunit (RPB1), RNA polymerase II second largest subunit (RPB2) and mitochondrial subunit (MtSU) gene regions (Bentley et al. 1998; Fourie et al. 2009; Maryani et al. 2019). The uniqueness of VCG 01212 to ECA proves that VCG testing remains important in Foc characterization, as it can provide information about the evolution and dissemination of pathogens.
References
Aguayo, J., Mostert, D., Fourrier-Jeandel, C., Cerf-Wendling, I., Hostachy, B., Viljoen, A., & Loos, R. (2017). Development of a hydrolysis probe-based real-time assay for the detection of tropical strains of Fusarium oxysporum f. sp. cubense race 4. PLoS One, 12, 1–20.
Beed, F. (2013). New banana disease to Africa find in Mozambique. CGIAR. http://www.rtb.cgiar.org/new-banana-disease-to-africa-found-in-mozambique/. Accessed 11 July 2019.
Bentley, S., Pegg, K., Moore, N., Davis, R., & Buddenhagen, I. (1998). Genetic variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense analyzed by DNA fingerprinting. Phytopathology, 88, 1283–1293.
Blomme, G., Ploetz, R., Jones, D. R., De Langhe, E., Price, N., Gold, C., Geering, A., Viljoen, A., Karamura, D., Pillay, M., Tinzaara, W., Teycheney, P. Y., Lepoint, P., Karamura, E., & Buddenhagen, I. (2013). A historical overview of the appearance and spread of Musa pests and pathogens on the African continent: Highlighting the importance of clean Musa planting materials and quarantine measures. Annals of Applied Biology, 162, 4–26.
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120.
Butler, D. (2013). Fungus threatens top banana. Nature, 504, 195–196.
Carter, B. A., Reeder, R., Byabachwezi, M. S. R., Kinyua, Z. M., Mbaka, J. N., Doyle, K., Nakato, V., Mwangi, M., Beed, F., Aritua, V., Lewis Ivey, M. L., Miller, S. A., & Smith, J. J. (2010). Identification of Xanthomonas vasicola (formerly X. campestris pv. musacearum), causative organism of banana Xanthomonas wilt, in Tanzania, Kenya and Burundi. Plant Pathology, 59, 403–403.
De Langhe, E., Karamura, D., & Mbwana, A. (2001). Tanzania Musa Expedition 2001. Rome: INIBAP/IPGRI.
Dita, M., Waalwijk, C., & Buddenhagen, I. (2010). A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen. Plant Pathology, 59, 348–357.
Edwards, M. C., & Gibbs, R. A. (1994). Multiplex PCR: Advantages, development, and applications. Genome Research, 3, 65–75.
Epstein, L., Kaur, S., Chang, P. L., Carrasquilla-Garcia, N., Lyu, G., Cook, D. R., Subbarao, K. V., & O’Donnell, K. (2017). Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology, 107, 463–473.
FAOSTAT. (2013). FAO statistical database. http://www.fao.org/faostat/en/#data/QC. Accessed 25 September 2017.
FAOSTAT. (2016). FAO statistical database. http://www.fao.org/faostat/en/#data/QC. Accessed 03 May 2019.
FAOSTAT. (2017). FAO statistical database. http://www.fao.org/faostat/en/#data/QC. Accessed 03 May 2019.
Fourie, G., Steenkamp, E. T., & Viljoen, A. (2009). Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Applied and Environmental Microbiology, 75, 4770–4781.
Gatsinzi, F., & Sebasigari, K. (1989). La maladie de Panama due à Fusarium oxysporum f. sp. cubense (E.F. Smith) Snyder & Hansen au sein de la communauté économique des pays des grands lacs (Burundi-Rwanda-Zaire). FAO Plant Protection Bulletin, 40, 68–74.
Geiser, D. M., Jiménez-Gasco, M. M., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T. J., Zhang, N., Kuldau, G. A., & O'Donnell, K. (2004). Fusarium-ID v. 1.0: A DNA sequence database for identifying Fusarium. European Journal of Plant Pathology, 110, 473–479.
Gold, C. S., Kiggundu, A., Abera, A. M. K., & Karamura, D. (2002). Diversity, distribution and farmer preferences of Musa cultivars in Uganda. Experimental Agriculture, 38, 39–50.
Gramaje, D., Pérez-Serrano, V., & Montes-Borrego, M. (2013). A comparison of real-time PCR protocols for the quantitative monitoring of asymptomatic olive infections by Verticillium dahliae pathotypes. Phytopathology, 103, 1058–1068.
Jiménez-Fernández, D., Montes-Borrego, M., & Jiménez-Díaz, R. (2011). In planta and soil quantification of Fusarium oxysporum f. sp. ciceris and evaluation of Fusarium wilt resistance in chickpea with a newly developed quantitative polymerase chain reaction assay. Phytopathology, 101, 250–262.
Kangire, A., Karamura, E. B., & Gold, C. (2000). Fusarium wilt of banana in Uganda, with special emphasis on wilt-like symptoms observed on east African highland cooking cultivars (Musa spp., AAA). Acta Horticulturae, 540, 343–353.
Karamura, D. A., Karamura, E. B., & Gold, C. S. (1996). Cultivar distribution in primary banana growing regions of Uganda. MusAfrica, 9, 3–5.
Karamura, E. B., Frison, E. A., Karamura, D. A., & Sharrock, S. (1998). Banana production systems in eastern and southern Africa. In C. Picq, E. Fourie, & E. A. Frison (Eds.), Bananas and food security (pp. 401–412). Montpellier: INIBAP.
Karangwa, P. (2015). Diversity and population structure of Fusarium oxysporum f.sp. cubense in East and Central Africa. Stellenbosch, South Africa: University of Stellenbosch, PhD thesis.
Karangwa, P., Blomme, G., Beed, F., Niyongere, C., & Viljoen, A. (2016). The distribution and incidence of banana Fusarium wilt in subsistence farming systems in east and Central Africa. Crop Protection, 84, 132–140.
Karangwa, P., Mostert, D., Ndayihanzamaso, P., Dubois, T., Niere, B. Z. F., Schouten, A., Blomme, G., Beed, F., & Viljoen, A. (2017). Genetic diversity of Fusarium oxysporum f. sp. cubense in east and Central Africa. Plant Disease, 102, 552–560.
Katoh, K., Kuma, K. I., & Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33, 511–518.
Kung’u, J. N., & Jeffries, P. (2001). Races and virulence of Fusarium oxysporum f. sp. cubense on local banana cultivars in Kenya. Annals of Applied Biology, 139, 343–349.
Leslie, J, F., & Summerell, B, A. (2006). The fusarium laboratory manual. 1st edition. Iowa, New York: Blackwell Publishing.
Lin, Y. H., Chang, J. Y., & Chang, P. F. L. (2009). Development of a molecular marker for specific detection of Fusarium oxysporum f. sp. cubense race 4. European Journal of Plant Pathology, 123, 353–365.
Lin, Y. H., Su, C. C., Chao, C. P., Chen, C. Y., Chang, C. J., Huang, J. W., & Chang, L. P. F. (2013). A molecular diagnosis method using real-time PCR for quantification and detection of Fusarium oxysporum f. sp. cubense race 4. European Journal of Plant Pathology, 135, 395–405.
Lodwig, E. M., Kung'u, J., & Jeffries, P. (1999). Molecular differences distinguish clonal lineages within east African populations of Fusarium oxysporum f. sp. cubense. Journal of Applied Microbiology, 86, 71–77.
Markoulatos, P., Siafakas, N., & Moncany, M. (2002). Multiplex polymerase chain reaction: A practical approach. Journal of Clinical Laboratory Analysis, 16, 47–51.
Maryani, N., Lombard, L., Poerba, Y. S., Subandiyah, S., Crous, P. W., & Kema, G. H. J. (2019). Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian Centre of origin. Study in Mycology, 92, 155–194.
Miller, S. A., Beed, F. D., & Harmon, C. L. (2009). Plant disease diagnostic capabilities and networks. Annual Review of Phytopathology, 47, 15–38.
Mostert, D., Molina, A. B., Daniells, J., Fourie, G., Hermanto, C., Chao, C. P., Fabregar, E., Sinohin, V. G., Masdek, N., Thangavelu, R., Li, C., Yi, G., Mostert, L., & Viljoen, A. (2017). The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS One, 12, 1–24.
Nkuba, J., Tinzaara, W., Night, G., Niko, N., Ndyetabula, I., Mukandala, L., Ndayihanzamaso, P., Niyongere, C., Gaidashova, S., Rwomushana, I., Opio, F., & Karamura, E. (2015). Adverse impact of Banana Xanthomonas wilt on farmers’ livelihoods in eastern and Central Africa. African Journal of Plant Science, 9, 279–286.
O’Donnell, K., Sutton, D. A., Rinaldi, M. G., Sarver, B. A. J., Balajee, S. A., Schroers, H. J., Summerbell, R. C., Robert, V. A. R. G., Crous, P. W., Zhang, N., Aoki, T., Jung, K., Park, J., Lee, Y. H., Kang, S., Park, B., & Geiser, D. M. (2010). Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology, 48, 3708–3718.
O'Donnell, K., Kistler, H. C., & Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. In: Proceedings of the National Academy of Science USA, 95(1998), 2044–2049.
Onyango, M., Karamura, D., Keeley, S., Manshardt, R., & Haymer, D. (2011). Morphological characterisation of east African AAB and AA dessert bananas (Musa spp.). International symposium on Banana: ISHS-ProMusa symposium on global perspectives on Asian challenges. Acta Horticulturae, 897, 95–105.
Perrier, X., Jenny, C., Bakry, F., Karamura, D., Kitavi, M., Dubois, C., Hervouet, C., Philippson, G., & De Langhe, E. (2018). East African diploid and triploid bananas: A genetic complex transported from South-East Asia. Annals of Botany, 20, 1–18.
Ploetz, R, C. (2000). Panama Disease: A classic and destructive disease of banana. Plant Health Progress, doi:https://doi.org/10.1094/PHP-2000-1204-01-HM
Ploetz, R. C. (2006). Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology, 96, 653–656.
Ploetz, R. C. (2015a). Fusarium wilt of banana. Phytopathology, 105, 1512–1521.
Ploetz, R. C. (2015b). Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Protection, 73, 7–15.
Ploetz, R. C., & Churchill, A. C. L. (2011). Fusarium wilt: The banana disease that refuses to go away. Proceedings of the international ISHS-ProMusa symposium on global perspectives on Asian challenges. Acta Horticulturae, 897, 519–526.
Ploetz, R. C., & Pegg, K. G. (2000). Fusarium wilt. In J. R. Jones (Ed.), Diseases of Banana, Abacá and Enset (pp. 143–159). New York: CABI Publishing.
Ploetz, R. C., Herbert, J., Sebasigari, K., Hernandez, J. H., Pegg, K. G., Ventura, J. A., & Mayato, L. S. (1990). Importance of Fusarium wilt in different banana-growing regions. In R. C. Ploetz (Ed.), Fusarium wilt of Banana (pp. 9–26). St Paul: APS Press.
Schrader, C., Schielke, A., & Johne, R. (2012). PCR inhibitors - occurrence, properties and removal. Journal of Applied Microbiology, 113, 1014–1026.
Stover, R, H. (1962). Fusarial Wilt (Panama disease) of Bananas and Other Musa Species. Kew: Commonwealth Mycological Institute.
Stover, R. H., & Buddenhagen, I. W. (1986). Banana breeding: Polyploidy, disease resistance and productivity. Fruits, 41, 175–191.
Tushemereirwe, W. K., Kangire, A., & Kubiriba, J. (2000). Fusarium wilt resistant bananas considered appropriate replacements for cultivars susceptible to the disease in Uganda. Uganda Journal of Agricultural Sciences, 5, 62–64.
Tushemereirwe, W. K., Kangire, A., & Kubiriba, J. (2004). Diseases threatening banana biodiversity in Uganda. African Crop Science, 12, 19–26.
Untergrasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., & Rozen, S. G. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Research, 40, 115–115.
Van Asten, P. J. A., Florent, D., & Apio, M. S. (2010). Opportunities and constraints for dried dessert banana (Musa spp.) export in Uganda. Acta Horticulturae, 879, 105–112.
Van Asten, P. J. A., Fermont, A. M., & Taulya, G. (2011). Drought is a major yield loss factor for rainfed east African highland banana. Agricultural Water Management, 98, 541–552.
Viljoen, A., Ma, L.-J., & Molina, A. B. (2019). Fusarium wilt (Panama disease) and monoculture banana production: Resurgence of a century-old disease. In A. Records & J. Ristaino (Eds.), Emerging plant diseases and global food security. St Paul: APS Press (In Press).
Wairegi, L., van Asten, P. J. A., Tenywa, M., & Bekunda, M. (2010). Abiotic constraints override biotic constraints in east African Highland Banana systems. Field Crops Research, 117, 146–153.
Zhang, X., Zhang, H., Pu, J., Qi, Y., Yu, Q., Xie, Y., & Peng, J. (2013). Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp. cubense tropical race 4 in soil. PLoS ONE, 8, e82841.
Acknowledgements
We would like to acknowledge Dr. Chunyu Li from the Institution of Fruit Tree Research, Guangdong Academy of Agricultural Sciences and Dr. Li-Jun Ma and Dr. Yong Zhang from University of Massachusetts, Amherst for bioinformatics support and insight on the selection of gene targets for primer design.
Funding
We would like to acknowledge funding provided by the International Institute of Tropical Agriculture (IITA) (ID: OPP1093845) through the project ‘Improvement of Banana for Smallholder Farmers in the Great Lakes Region of Africa). Additional funding support was provided by the Belgian Directorate General for Development Cooperation and Humanitarian Aid (DGD) through the Consortium for Improving Agricultural Livelihoods in Central Africa (CIALCA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This research fully complies with Ethical Standards applicable for this journal and the relevant national and international ethics standards and professional codes of conduct.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ndayihanzamaso, P., Karangwa, P., Mostert, D. et al. The development of a multiplex PCR assay for the detection of Fusarium oxysporum f. sp. cubense lineage VI strains in East and Central Africa. Eur J Plant Pathol 158, 495–509 (2020). https://doi.org/10.1007/s10658-020-02092-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-02092-9