Abstract
The aim of the present study was to determine the virulence structure of powdery mildew of oats (Blumeria graminis DC.f. sp. avena) in Poland in the years 2010–2013. For this purpose, powdery mildew isolates were collected from three experimental stations in Poland. To assess the virulence of the isolates, eight oat varieties with different responses to the pathogen were used. The results showed that a significant proportion of powdery mildew isolates found in Poland overcame the resistance genes of varieties Bruno (Pm6), Jumbo (Pm1) and Mostyn (Pm3). In contrast, lines Av1860 (Pm4), Am27 (Pm5) and Cc3678 (Pm2) were completely resistant to all pathogen isolates involved in the experiment. Changes constantly occurring in the powdery mildew population perfectly reflect diversity indexes, which were the smallest in the first year of observation, where in the following years these parameters were significantly higher. It is worth noting that the presence of powdery mildew is seasonal and local, which is reflected in the prevalence of the disease in a defined area of the country.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In principle, fungal diseases are the primary limiting factor in the cultivation of oats (Avena sativa L.) in many countries. One of the most common diseases in agricultural fields is powdery mildew. The disease is caused by parasitic fungus Blumeria graminis DC. f. sp. avenae Em. Marchal, which has a very effective spreading mechanism that is difficult to control through the application of treatments, such as crop rotation (Roderick et al. 2000; Stevens et al. 2004). The most effective and environmentally friendly method of controlling and limiting the consequences of infection is the introduction of crop cultivars containing effective resistance genes (Okoń et al. 2016).
The problem of annually occurring powdery mildew on crops particularly concerns cooler, humid regions of cultivations such as maritime north-western Europe, North America (Aung et al. 1977; Schwarzbach and Smith 1988), and the countries of Eastern Europe, including Poland (Sebesta et al. 1991). In recent years, an increasing number of reports have been published on the appearance of new races (Erysiphales) in Poland (Świderska et al. 2005; Wołczańska and Mułenko 2002). First symptoms of disease are grey-white colonies of cottony mycelia on the leaf surface that spread and form a dense mat. As an obligate parasite, the fungus depletes the host of nutrients, resulting in chlorosis and necrosis of infected and uninfected tissues and reduced vegetative and reproductive growth. Early and continued infection has most severe effects on tillering, inflorescence size and consequently seed number and size, whereas later infections, particularly on the upper leaves, affect grain filling (Clifford 2012). The losses caused by powdery mildew are significant, ranging from 5 to 10%, and even up to 30% in western Europe (Hsam et al. 1997; Zhang et al. 2005)
The interaction of the host and powdery mildew is closely related to the ‘gene-for-gene’ hypothesis, which implies that the virulence gene (Avr) in the genome of the pathogen is directed against the resistance gene (R) in a plant (Flor 1971; Heath 2000). In oats, so far eight genes of resistance to powdery mildew have been identified and described (Hsam et al. 2014; Jones and Jones 1979). The aim of the study was to determine the virulence of powdery mildew isolates obtained from breeding stations in Poland to the known major resistance genes of oats.
Materials and methods
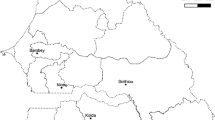
Oat leaf fragments with visible symptoms of powdery mildew were collected from the following locations: Choryń, Czesławice and Strzelce across 2010–2013. Geographical origin of isolates and years of occurrence are presented in Fig. 1. At least, 20 single-spore isolates were established under laboratory conditions from each sampled powdery mildew population according to the method described by Okoń and Kowalczyk (2012).
In order to check the degree of virulence, host-pathogen tests were carried out using 8 oat genotypes with different resistance genes to powdery mildew (Table 1). Cultivar Fuchs without any resistance gene to powdery mildew was used as susceptible control.
Host-pathogen tests were carried out on the first leaves of 10-day-old seedlings. Leaf segments were placed into 12-well culture plates with 6 g/l agar and 35 mg/l benzimidazole. The plates with the leaf segments were inoculated in a settling tower by spreading 500–700 spores of powdery mildew per 1 cm2. Then the plates were incubated in a phytotron at 17 °C and illuminance of approximately 4 kLx.
Resistance reactions were determined ten days after inoculation and scored according to a 0–4 modified scale (Mains 1934): where 0 = no infection, no visible symptoms; 1 = highly resistant, fungal development limited, no sporulation; 2 = moderately resistant, moderate mycelium with some sporulation; 3 = moderately susceptible, extensive mycelium, more sporulation; 4 = highly susceptible, large colonies, abundant sporulation. If disease symptoms were scored as 0, 1 or 2, the isolates were classified as avirulent to known genes against oat powdery mildew. If disease symptoms were scored as 3 or 4, the isolates were classified as virulent.
Descriptive statistics were calculated using the spreadsheet HaGiS Tool (Herrmann et al. 1999). For each year and sampling site calculated the following population parameters:
-
I.
Virulence frequency (p) as p = x/n;
-
II.
Complexity;
-
III.
Gilmour codes:
-
IV.
Diversity indexes such as:
-
Simple Index (Q) as Q = R/N;
-
Shannon Index (H′) as H ' = − ∑p i ln (p i );
-
Gleason Index (H g ) as H g = (r – 1)/ log e (N);
-
Simpson Index (λ) as λ = ∑\( {p}_i^2 \);
-
V.
Evenness (J H’ ) as \( {J}_{H^{\hbox{'}}} \)= H '/H max, where H max = ln S.
Results
One of the basic values for this type of analysis, virulence frequency, was calculated on the basis of host-pathogen tests conducted on genotypes with defined resistance genes to oat powdery mildew (Table 1). The frequency of virulence (p) was calculated for each differential, where x is the number of times a virulent reaction type (those which causing reaction types 3 or 4) was detected and n is the total number of samples tested in a particular year (Table 2).
In 2010, symptoms of powdery mildew on oats were observed in the western part of the country. Leaves of diseased plants were collected in experimental plots of the breeding station in Choryń (Wielkopolska Province). All single-spore isolates derived from populations collected in Choryń were characterized by a complete virulence (100%) to the resistance genes Pm6, Pm1 and Pm3. However, isolates did not overcome the resistance determined by Pm4, Pm5 and Pm2 genes (0%). Virulence against the Pm7 gene remained at the level of 5%, which also indicated a high degree of resistance conditioned by this gene.
In 2011, as in the previous year, disease symptoms were observed in the western part of the country. Populations of the pathogen were also collected from experimental plots of the breeding station in Choryń. In 2011 a decrease in virulence frequency was recorded (Fig. 3) against Pm1, Pm3 (5%) and Pm6 (20%) compared to 2010. Virulence of isolates for genes Pm2, Pm4, Pm5 and Pm7 has not changed in 2011.
In 2012, symptoms of powdery mildew of oats were only observed in central Poland. Populations of the pathogen were obtained from the experimental station in Strzelce (Łódź Province). The isolates obtained were characterized by a 5% decrease in the virulence frequency for genes Pm1 and Pm3 compared to previous years. In comparison to the previous year, virulence frequency against the Pm6 gene has increased by 10%, but was lower than in 2010. Similarly as in previous years, virulence frequency against genes Pm2, Pm4, Pm5 and Pm7 was at the same level.
In 2013, powdery mildew on oats occurred in the central and eastern part of the country. Pathogen isolates were established from populations collected in Strzelce (Łódź Province) and Czesławice (Lublin Province). Despite the increased number of isolates tested, the results were similar to previous years. A slightly lower percentage of isolates was found to break the resistance conditioned by genes Pm6 and Pm1 (5%), while a slight increase in frequency (5%) was observed for the Pm3 gene. In 2013, no virulent isolates were observed for Pm7. The isolates obtained in 2013 were again avirulent for Pm2, Pm4 and Pm5 (Table 2).
Isolate complexity is another parameter characterizing the pathogen population. This parameter specifies the number of varieties (X-axis) that have been infected by a given percentage of isolates (Y-axis) with respect to particular years (Fig. 2). Powdery mildew isolates analyzed in the present study generally broke the resistance of three out of seven resistance genes used in the host-pathogen tests; these were Pm1, Pm3 and Pm6 genes. In 2010, more than 90% of the analyzed isolates of powdery mildew overcame the resistance of these genes, while in subsequent years, the number of isolates reached 70–80%. In 2010, powdery mildew isolates were virulent to a minimum of three and a maximum of four genes. In 2011 and 2012, the obtained powdery mildew isolates were virulent to a minimum of one and a maximum of four genes. Isolates collected in 2013 were virulent to a minimum of one and a maximum of three genes. None of the powdery mildew isolates was able to overcome the resistance of all analyzed resistance genes (Fig. 2).
Virulence frequency of powdery mildew isolates obtained from the same location but in different years was similar (Fig. 3). This indicates a low change in the population of powdery mildew at yearly intervals.
Pathotypes of isolates were determined using a three-digit code developed by Gilmour (1973) based on the model of infection of individual control lines by the tested isolates of powdery mildew (Table 3). Fig. 4 shows the distribution of individual pathotypes of the analyzed powdery mildew isolates. Pathotype 700 was the most common pathotype, and was represented by approx. 80% of the analyzed oat powdery mildew isolates. Pathotype 200 was the least common pathotype in experiment and was represented by 2% of the analyzed isolates. In 2010, two pathotypes were identified, five in 2011, while six powdery mildew pathotypes were recorded in 2012 and 2013 (Table 4). Pathotype variability was very low due to the small number of available control forms and due to the fact that almost all isolates were virulent to genes Pm1, Pm3 and Pm6.
Table 4 contains widely used indexes describing diversity of the studied powdery mildew populations. The simplest algorithm of calculation diversity is Simple Index (Q), where R is the number of distinct races (pathotypes) detected in the sample and N is the total number of isolates in the sample. The lowest value of Simple Index characterized isolates was from Choryń in 2010 (0.10), the highest value was in Strzelce in 2012 (0.30).
The genetic diversity of virulence factors was calculated by the Shannon’s information function (H′) (Shannon and Weaver 1949), where p i is the relative frequency of occurrence of virulence factors in the pathogen population sampled. As a result, the most biodiverse powdery mildew population was from Choryń in 2011 (0.98), the population with the smallest biodiversity characterized was also from Choryń but in 2010 (0.20).
The Gleason Index (H g ) reflects the number of races per sample, in which r is the number of races in the sample and N is the total number of isolates in the sample. Thus, the Gleason Index recognizes that the probability of detecting a new race with each new isolate declines as the total number of isolates in the sample increase. This index ranged between 0.33 in Choryń in 2010 and 1.67 in Strzelce in 2012.
Simpson’s Index of diversity ‘λ’ (Simpson 1949), which indicates the dominance or concentration of virulence factors in the population. This index ranged between 0.1 in Choryń in 2010 and 0.51 in Choryń in 2011.
The index of evenness, \( {J}_{H^{\hbox{'}}} \) is used to confront empirical and maximum theoretical values of H ', and S = maximum number of virulent factors in the pathogen population (Sheldon 1969). Pathotype evennes in this study ranged between 0.29 in Choryń in 2010 and 0.61 in Choryń in 2011.
Discussion
The experiments carried out in the present study are the first that concern the analysis of virulence frequency of oat powdery mildew in Poland. Blumeria graminis DC. f. sp. avenae is a fungus whose conidia are transported with the wind. In Poland, most of the winds come from the west, which in conjunction with the fact that the vegetation in western Europe begins earlier than in Poland, results in the common occurrence of this fungus in the spring on cereal plantations in our country (Limpert et al. 1999, 2000). In addition, weather conditions observed in recent years favored the occurrence and spread of powdery mildew of cereals. Observations carried out in the present study confirm the spread of the disease from the West to the East of the country. In 2010 and 2011, the signs of powdery mildew were observed only in the western part of the country. In subsequent years of research, signs of powdery mildew were detected in central Poland (2012) and in eastern Poland (2013).
Diversity indices provide important information about rarity and commonness of species in a community. The ability to quantify diversity in this way is an important tool for biologists trying to understand population structure. Based on the own results and the results of other authors we can conclude that the diversity in the population of powdery mildew from year to year is increasing. Czembor et al. (2014) in their study analyzed virulence structure of the powdery mildew population occurring on triticale in Poland. They used twenty-one wheat and seven triticale differentials. The diversity indexes, both for wheat and triticale were the lowest in the first year of the experiment. In subsequent years these values were higher. For example Czembor et al. (2014) showed that Shannon’s Index (H′) of the tested wheat population amounted to 5.60 in 2008 while in 2009 and 2010 were 5.99 and 5.90 respectively. In the present study the smallest diversity indexes were also recorded in the first year of observation (in 2010 Q = 0.10; H g = 0.33; H′ = 0.20), the following years were characterized by higher rates (e.g. in 2012 Q = 0.30; H g = 1.67; H′ = 0.96).
Mutations and sexual reproduction are considered as the main source of genetic variation in powdery mildew. Referring to the example of B. graminis f. sp. hordei causal for the occurrence of powdery mildew in barley, the rate of spontaneous mutations in a single locus from avirulence to virulence ranges from 10−9 to 10−6 per generation of the pathogen (Marshal 1977; Torp and Jensen 1985). These factors lead to the emergence of virulent pathotypes that systematically break the resistance conditioned by the resistance genes present in oats cultivars. Thus far, the oat breeding programs have used Pm1, Pm3 and Pm6 genes (Hsam et al. 1997, 1998; Okoń 2012). However, resistance conditioned by these genes has already been broken by the existing races of the pathogen (Okoń 2015). In the present study, the highest level of virulence was observed for these genes among the analyzed powdery mildew isolates; it ranged from 80 to 100%.
The current study also used oat lines carrying genes Pm2, Pm4, Pm5 and Pm7 to estimate the virulence of the population of powdery mildew present in Poland in the years 2010–2013. Hsam et al. (1997) have identified the APR122 line with the Pm7 gene, demonstrating a high level of resistance to powdery mildew isolates used by these authors. None of the powdery mildew isolates used by Hsam et al. (1997, 1998) overcame the resistance of this gene. Research conducted by Okoń (2015) confirmed the high level of resistance conditioned by the Pm7 gene. In the present study, the level of virulence of the analyzed powdery mildew isolates for this gene was 5%. This demonstrates the high level of resistance conditioned by this gene. It can also be assumed that this resistance will persist in the coming years.
Oat genotypes carrying the Pm4 gene were characterized as highly resistant to powdery mildew (Hsam et al. 1997, 1998, 2014). In the present study, none of the powdery mildew isolates have overcome the resistance conditioned by this gene. Numerous studies have shown that this gene has not been used so far in oat breeding programs (Hsam et al. 1997, 1998; Okoń 2012). The Pm5 gene was introduced into A. sativa from A. macrostachya (Yu and Herrmann 2006). The experiments conducted in this study showed that this gene is highly resistant to powdery mildew isolates found in Poland in the years 2010–2013. The level of virulence for this gene during all analyzed years was equal to 0. The same was true for the Pm2 gene. However, the available sources of literature do not provide information on the use of these genes in oat breeding programs. Lack of virulence of the analyzed powdery mildew isolates for genes Pm2, Pm4 and Pm5, and a very low level of virulence for the Pm7 gene indicate the great potential of these genes for cultivar improvement. Absence of changes in the level of virulence of the analyzed powdery mildew isolates obtained in 2010–2013 from different regions of the country also indicates that the resistance conditioned by these genes can persist for a long period of time before a race of pathogen occurs that will be able to overcome this resistance.
References
Aung, T., Thomas, H., & Jones, I. T. (1977). The transfer of the gene for mildew resistance from Avena barbata (4x) into the cultivated oat A. sativa by an induced translocation. Euphytica, 26, 623–632. doi:10.1007/BF00021687.
Clifford, B. C. (2012). Diseases, pests and disorders of oats. In R. Welch (Ed.), The oat crop: Production and utilization (pp. 252–276). London: Chapman & Hall.
Czembor, H. J., Domeradzka, O., Czembor, J. H., & Mańkowski, D. R. (2014). Virulence structure of the powdery mildew ( Blumeria graminis ) population occurring on triticale (X Triticosecale ) in Poland. Journal of Phytopathology, 162(7–8), 499–512. doi:10.1111/jph.12225.
Flor, H. H. (1971). Current status of the Gene-for-Gene concept. Annual Review of Phytopathology, 9, 275–296. doi:10.1146/annurev.py.09.090171.001423.
Gilmour, J. (1973). Octal notation for designing physiologic races of plant pathogens. Nature, 242, 620.
Heath, M. C. (2000). Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology, 3, 315–319. doi:10.1016/S1369-5266(00)00087-X.
Herrmann, A., Löwer, C. F., & Schachtel, G. A. (1999). A new tool for entry and analysis of virulence data for plant pathogens. Plant Pathology, 48, 154–158. doi:10.1046/j.1365-3059.1999.00325.x.
Hsam, S. L. K., Mohler, V., & Zeller, F. J. (2014). The genetics of resistance to powdery mildew in cultivated oats (Avena sativa L.): Current status of major genes. Journal of Applied Genetics, 55, 155–162. doi:10.1007/s13353-014-0196-y.
Hsam, S. L. K., Pederina, E., Gorde, S., & Zeller, F. J. (1998). Genetic studies of powdery mildew resistance in common oat (Avena staiva L.). II. Cultivars and breeding lines grown in northern and Eastern Europe. Hereditas, 129, 227–230.
Hsam, S. L. K., Peters, N., Paderina, E. V., Felsenstein, F., Oppitz, K., & Zeller, F. J. (1997). Genetic studies of powdery mildew resistance in common oat (Avena sativa L.) I. Cultivars and breeding lines grown in Western Europe and North America. Euphytica, 96, 421–427. doi:10.1023/A:1003057505151.
Jones, I.T., & Jones, E.R.L. (1979). Mildew of oats. UK cereal pathogen virulence survey 1978. Annual Rep. 59-63.
Limpert, E., Bartos, P., Graber, W. K., Müller, & Fuchs, J. G. (2000). Increase of virulence complexity of nomadic airborne pathogens from west to east across Europe. Acta Phytopathol. Entomologica Hungarica, 35, 261–272.
Limpert, E., Godet, F., & Muller, K. (1999). Dispersal of cereal mildews across Europe. Agric. Forest Meteorol., 97, 293–308.
Mains, E. B. (1934). Inheritance of resistance to powdery mildew, Erysiphe graminis tritici, in wheat. Phytopathology, 24, 1257–1261.
Marshal, D. R. (1977). The advantages and hazards of genetic homogenetity. Annals of the New York Academy of Sciences, 287, 1–20.
Okoń, S. M. (2012). Identification of powdery mildew resistance genes in polish common oat (avena sativa l.) cultivars using host-pathogen tests. Acta Agrobotanica, 65, 63–68. doi:10.5586/aa.2012.008.
Okoń, S. M. (2015). Effectiveness of resistance genes to powdery mildew in oat. Crop Protection, 74, 48–50. doi:10.1016/j.cropro.2015.04.004.
Okoń, S., & Kowalczyk, K. (2012). Deriving isolates of powdery mildew (Blumeria graminis DC. F.Sp. avenae Em. Marchal.) in common oat (Avena sativa L.) and using them to identify selected resistance genes. Acta Agrobotanica, 65, 155–160. doi:10.5586/aa.2012.069.
Okoń, S., Paczos-Grzęda, E., Ociepa, T., Koroluk, A., Sowa, S., Kowalczyk, K., & Chrząstek, M. (2016). Avena sterilis L. Genotypes as a potential source of Resistance to oat powdery mildew. Plant Dis. 100, 2145–2151. doi:10.1094/PDIS-11-15-1365-RE.
Roderick, H. W., Jones, E. R. L., & Sebesta, J. (2000). Resistance to oat powdery mildew in Britain and Europe: A review. The Annals of Applied Biology, 136, 85–91.
Schwarzbach, E., & Smith, I. M. (1988). Erysiphe graminis DC. In I. M. Smith, J. Dunez, R. A. Lelliot, D. H. Philips, & S. A. Archer (Eds.), European handbook of plant diseases. Oxford: Blackwell.
Sebesta, J., Kummer, M., Roderick, H. W., Hoppe, H. D., Cervenka, J., Swierczewski, A., & Muller, K. (1991). Breeding oats for resistance to rusts and powdery mildew in central Europe. Ochr. Rostl., 27, 229–238.
Shannon, C. E., & Weaver, W. (1949). The mathematical theory of communication. Urbana: University of Illinois Press.
Sheldon, A. L. (1969). Equitability indices: Dependence on the species count. Ecology, 50, 466–467.
Simpson, E. H. (1949). Measurement of diversity. Nature, London, 163, 688.
Stevens, E.J., Armstrong, K.W., Bezar, H.J., Griffin, W.B., & Hampton, J.G. (2004). Fodder oats : An overview, in: Suttie, J.M., Reynolds, S.G. (Eds.), Fodder Oats: An Overview. Rome, pp. 11–18.
Świderska, U., Wołczańska, A., Kozłowska, M., Mułenko, W., & Mamczarz, M. (2005). Recent collections of powdery mildew in Poland. Acta Mycologica, 40, 49–61.
Torp, J., & Jensen, H. P. (1985). Screening for spontaneous virulent mutantes to Erysiphe graminis DC f.Sp. hordei on barley lines with resistance genes Ml-a1, Ml-a6, Ml-a12 and Ml-g. Phytopathologische Zeitschrift, 112, 17–27.
Wołczańska, A., & Mułenko, W. (2002). New collections of powdery Mildews (Erysiphales ) in Poland. Polish Bot. J., 47, 215–222.
Yu, J., & Herrmann, M. (2006). Inheritance and mapping of a powdery mildew resistance gene introgressed from Avena macrostachya in cultivated oat. Theoretical and Applied Genetics, 113, 429–437.
Zhang, Z., Henderson, C., Perfect, E., Carver, T. L. W., Thomas, B. J., Skamnioti, P., & Gurr, S. J. (2005). Of genes and genomes, needles and haystacks : Blumeria graminis and functionality. Molecular Plant Pathology, 6, 561–575. doi:10.1111/J.1364-3703.2005.00303.X.
Acknowledgements
This research was financed by the Ministry of Agriculture and Rural Development in the frame of basic research program for biological progress in crop production under the title: “Identification and localization of DNA markers for selected powdery mildew resistance genes in common oat and pyramiding of the effective resistance genes in the oat genome”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Okoń, S.M., Ociepa, T. Virulence structure of the Blumeria graminis DC.f. sp. avenae populations occurring in Poland across 2010–2013. Eur J Plant Pathol 149, 711–718 (2017). https://doi.org/10.1007/s10658-017-1220-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1220-y