Abstract
Recognition of non-self in plants is mediated by specialised receptors that upon pathogen perception trigger induction of host defence responses. Primary, or basal, defence is mainly triggered by trans-membrane receptors that recognise conserved molecules released by a variety of (unrelated) microbes. Pathogens can overcome these basal defences by the secretion of specific effectors. Subsequent recognition of these effectors by specialised receptors (called resistance proteins) triggers induction of a second layer of plant defence responses. These responses are qualitatively similar to primary defence responses; however, they are generally faster and stronger. Here we give an overview of the predicted (domain) structures of resistance proteins and their proposed mode of action as molecular switches of plant innate immunity. We also highlight recent advances revealing that some of these proteins act in the plant nucleus as transcriptional co-regulators and that crosstalk can occur between members of different resistance protein families.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to distinguish self from non-self is the most fundamental aspect of an immune system. Recognition of invaders in both plants and animals is mediated by extra- and intracellular immune receptors. Unlike vertebrates, which have adaptive molecular receivers in specialised mobile cells, plants rely on a spectrum of predetermined receptors expressed in non-mobile cells. Therefore, in plants pathogen-arrest is orchestrated by the cells encountering the pathogen and the systemic signals that originate from these cells.
It has been hypothesized that early land plants contained trans-membrane receptors at their cell surface capable of recognizing microbe- or pathogen-associated molecular patterns (MAMPS or PAMPS) such as cell wall fragments, chitin or peptide motifs in bacterial flagella (Ausubel 2005; Chisholm et al. 2006; Nürnberger and Kemmerling 2006). Recognition of these common and slowly evolving PAMPs triggers the induction of the primary or basal defence responses, nowadays also referred to as PTI (PAMP-triggered immunity; Jones and Dangl 2006). Evolution of this ancient immune system put a constraint on pathogenic microbial populations as it limited their host range and it forced them to develop counter strategies to overcome PTI. This selection pressure has likely to have resulted in the acquisition of virulence effector proteins that suppress basal plant defence. Many plant pathogens have been shown to produce, and deliver, effector proteins in the host (Birch et al. 2006; Catanzariti et al. 2007; Grant et al. 2006; Jones and Dangl 2006). In the subsequent evolutionary struggle to combat these pathogens plants evolved means to recognise the secreted effector proteins and to mount a robust amplified defence response. This type of secondary defence is referred to as effector-triggered immunity (ETI) and is mediated by resistance (R) proteins. In broad terms the defence responses associated with both PTI and ETI are qualitatively similar; however, those associated with the latter are generally faster and stronger and are often accompanied by localized cell death around the infection site (Jones and Dangl 2006). Although what actually stops pathogen proliferation is still unclear in most cases, new data has recently become available on the receptors that switch on defence after pathogen recognition. This review aims to provide a current overview of the structure and function of these R proteins and highlights recent advances.
Resistance proteins
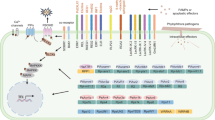
A common feature of receptors involved in pathogen perception is the leucine-rich repeat (LRR) domain (Fig. 1). This domain is present both in PAMP receptors, where it is fused to a transmembrane domain and a cytoplasmic kinase domain [receptor-like kinase (RLK)], and in the majority of R proteins (Nürnberger and Kemmerling 2006). Some R proteins structurally resemble the PAMP RLK receptors, such as the rice Xa21 and Xa26 proteins (Sun et al. 2004). In others, like the tomato Cladosporium fulvum resistance (Cf) proteins (Rivas and Thomas 2005), the extracellularly localized LRR is also fused to a transmembrane domain. However, in these proteins [receptor-like protein (RLP)] no recognisable intracellular signalling domains can be discerned (Rivas and Thomas 2005).
Schematic representation of domains found in plant LRR R proteins. Domains are not drawn to scale. TIR Toll/interleukin-1 receptor, CC coiled coil, NB nucleotide binding, ARC1/2 APAF1, R protein and CED4, LRR leucine rich repeat, SD solanaceous domain, BED BEAF/DREAF zinc finger domain, TM transmembrane, Kin kinase, WRKY WRKY transcription factor
In the majority of currently identified R proteins, however, the LRR resides in the cell and is fused to a nucleotide binding (NB) domain. The core nucleotide binding fold in these proteins is part of a larger entity called the NB-ARC domain due to its presence in Apaf-1 (apoptotic protease-activating factor-1), R proteins and CED-4 (Caenorhabditis elegans death-4 protein; van der Biezen and Jones 1998). Database searches have revealed a structurally related domain in animal proteins named NACHT (NAIP, CIITA, HET-E, and TP1) or NOD (for nucleotide-oligomerisation domain) domain (Leipe et al. 2004; Rairdan and Moffett 2007; Ting et al. 2006). Many of these proteins act as receptors sensing intracellular perturbations, such as the presence of microbial compounds (e.g. MAMP recognition by NACHT-LRRs), or cytochrome c leaking from mitochondria (e.g. Apaf-1). Like in R proteins, the NB-ARC/NACHT/NOD domains in these proteins are fused to a repeat structure such as an LRR or WD40 repeat domain (Leipe et al. 2004). Although these proteins share similar mechanistic and structural features, they appear to have evolved independently (Ausubel 2005; Leipe et al. 2004). The ubiquitous use of fused NB-repeat structures throughout the plant and animal kingdom probably reflects the biochemical suitability of such a module for coupled ligand recognition and subsequent activation of downstream signal transduction.
The NB-LRR core of plant R proteins is often equipped with variable amino- and sometimes also carboxy-terminal domains. Figure 1 gives an overview of the various structural domain decorations found in the different subfamilies of NB-LRR R proteins. The two major NB-LRR subfamilies are distinguished by either the presence or absence of an amino-terminal Toll/interleukin-1 receptor-like domain (TIR; Meyers et al. 1999). As many non-TIR NB-LRR proteins contain predicted coiled coil (CC) motifs, this family is collectively referred to as CC-NB-LRRs.
The TIR-NB-LRR (TNL) and CC-NB-LRR (CNL) members do not only differ in their amino-terminal extensions but also in certain motifs in their NB domains, thereby separating them into two evolutionary divergent classes (see below; Meyers et al. 1999; Pan et al. 2000). Members of the CNL group can be further divided based on the presence of additional domains at their amino-terminus. One example is a long extension, which has so far only been found in the Solanaceae and is therefore referred to as solanaceous domain (SD; Mucyn et al. 2006; Rairdan and Moffett 2007). Another example is the BED (named after the BEAF and DREF proteins) zinc-finger DNA- binding domain found in rice Xa1 and in NB-LRRs of poplar (Aravind 2000; Tuskan et al. 2006). At the carboxy-terminus flanking the LRR some R proteins carry extensions without recognizable domains, an exception being the Arabidopsis RRS-1-R protein that contains a typical WRKY DNA-binding domain at its carboxy-terminus (Deslandes et al. 2003).
Characteristic features of R protein domain structures
The class of extracellular RLP R proteins, founded by the Cf proteins, has mainly been found in Solanaceous species. These consist mainly of an extracellular LRR domain anchored in the plasma membrane (Rivas and Thomas 2005). Recently, in rice a new type of RLK R protein was identified: Pi-d2. In Pi-d2, which confers resistance to rice blast, the LRR is replaced by a B-lectin domain (lecRLK). Similar to RLPs and RLKs the extracellular domain is proposed to be involved in detection of the pathogen (Chen et al. 2006).
The intracellular NB-LRR R proteins are numerous and are present in large gene families in Arabidopsis (∼150), rice (∼400) and poplar (∼400) (Meyers et al. 2003; Monosi et al. 2004; Tuskan et al. 2006). They are among the largest proteins found in plants and range from 860–1900 amino acid residues in size (McHale et al. 2006). As mentioned before, these NB-LRRs often contain four domains connected by linkers; a variable N-terminus, the NB-ARC domain, the LRR domain and a variable C-terminal extension. Unfortunately, so far, no crystal structure has been determined for any plant R protein or parts thereof. However, 3D modelling templates are available for the LRR and NB-ARC domains to predict their structure (Albrecht and Takken 2006; McHale et al. 2006; Takken et al. 2006). Specific features of each domain will be discussed below.
The LRR domain
The LRR domain represents the major part of RLP R proteins and is composed of a variable number of repeats fitting the 24-amino acid residue consensus motif LxxLxxLxLxxNxLxGxIPxxLGx (L, leucine; x, any amino acid; N, asparagine; G, glycine; I, isoleucine; P, proline; Kajava 1998). In most RLPs this extracellular LRR domain is interrupted by a spacer region not fitting the consensus sequence, thereby dividing the LRR into three subdomains. The largest, amino-terminal, part consists of 21–28 hyper-variable repeats, the middle part represents the spacer and the remainder consists of three or four relatively conserved LRRs (Rivas and Thomas 2005). In RLK R proteins a division of the LRR domain into sub-domains has not been observed (Sun et al. 2006).
Unlike the LRR in RLPs and RLKs, the LRR in NB-LRR R proteins fits a shorter consensus motif that consists of 14 residues [LxxLxxLxLxxC/Nxx (C, cysteine; other symbols as above)] embedded in a repeat with a typical length of 24–28 residues (Kajava 1998). Based on crystal structures of non-plant LRRs the 14-residue core is predicted to form a β-sheet and an attached loop region. The remaining part of the repeat forms a spacer allowing the β-sheets to stack, thereby forming a large right-handed super helical β-sheet (Kobe and Deisenhofer 1994). So far the crystal structures of two plant LRRs, polygalacturonase-inhibiting protein-2 (PGIP2; containing an extracellular LRR), and the cytoplasmic TIR-1 auxin receptor have been elucidated (Di Matteo et al. 2003; Tan et al. 2007). These structures revealed differences with non-plant LRRs. The PGIP2-LRR has two β-sheets in each repeat, connecting the first with an α-helix in the spacer, resulting in an extended and slightly curved super-structure (Di Matteo et al. 2003). The LRR of TIR-1 forms a horse shoe-like structure in which the β-strands lie at the concave side, whereas the mainly α-helical spacers lie at the convex side. In contrast to other crystallised LRR proteins, the TIR-1 LRR has a cofactor, the inositol-6-phosphate (InsP6), tightly bound in the middle of the horseshoe that provides a ‘floor’ for the auxin-binding pocket. Surprisingly, the auxin-binding interface is not formed by residues embedded in the β-sheet, but by three intra-repeat loops that stick out of the plane of the horseshoe (Tan et al. 2007). It will be interesting to determine whether the LRRs of NB-LRR proteins adopt a similar structure that is distinct from that of non-plant LRRs. The LRR domain structure is perfectly suited to mediate protein-protein interactions and ligand binding (Kobe and Deisenhofer 1994). Basically two different types of R protein LRR classes can be discerned, those with high genetic diversity and the others showing little variation. This difference has been proposed to reflect the recognition mechanism of the pathogen’s effector, direct versus indirect (Ellis et al. 2007). No effector protein, however, has yet been identified that directly binds the LRR. The LRR interactors identified are chaperones that might be required for proper folding of the LRR domain (Azevedo et al. 2006; Bieri et al. 2004; Holt et al. 2005; Takahashi et al. 2003). These chaperones include heat-shock proteins such as HSP90 and HSP17 and co-chaperones such as protein phosphatase 5, SGT1 and RAR1 (De la Fuente van Bentem et al. 2005; Hubert et al. 2003; Liu et al. 2004; Liu et al. 2002; Takahashi et al. 2003). Interestingly, the LRR domain appears to interact with the N-terminal part of NB-LRR proteins as exemplified by Rx, Bs2 and N (Leister et al. 2005; Moffett et al. 2002; Ueda et al. 2006). As discussed below, these intramolecular interactions are probably important for the regulation of NB-LRR protein activity and thus for the induction of ETI.
The amino-terminus of plant NB-LRR proteins
In animals, both CC and TIR domains have been implicated in protein-protein interactions, which is also predicted to occur in plant NB-LRR proteins as they are thought to interact with domain-specific downstream signalling components (Feys and Parker 2000). Recent observations indicate that the amino-terminus of at least some NB-LRR proteins also binds host proteins that are subject to attack by pathogen effectors (so-called virulence targets or guardees) in order to guard them and monitor their perturbations. Observations supporting the role of the amino terminus in this guard function are the binding of RIN4 to the R proteins RPM1 and RPS2, PBS1 kinase binding to RPS5, NIP1(N-interacting protein 1) binding to N and Pto binding to Prf (Ade et al. 2007; Burch-Smith et al. 2007; Dinseh-Kumar, personal communication; Mackey et al. 2002; Mucyn et al. 2006). These virulence targets are either cleaved (RIN4 and PBS-1), phosphorylated (RIN4) or modified in an unknown way (NIP1 and Pto) by their attacking effector proteins (AvrRpm1/AvrB and AvrRpt2 for RIN4, AvrPphB for PBS-1, P50 for NIP1 and AvrPto/AvrPtoB for Pto; Axtell et al. 2003; Burch-Smith et al. 2007; Mackey et al. 2002; Mucyn et al. 2006; Shao et al. 2003; Dinesh-Kumar, personal communication). The modification of the guardees is believed to trigger the guarding NB-LRR protein to activate ETI. In the case of R proteins that directly recognise pathogen effectors it is not known whether the effectors also bind to the N-terminal domain or whether they bind to other domains.
Besides the observed interactions with guardees, the N-terminal domain can be involved in homotypic TIR-TIR interactions as shown for the tobacco R protein N resulting in NB-LRR oligomerisation upon activation (Mestre and Baulcombe 2006; see below).
The overall 3D structure of plant TIR domains is not known. However, crystal structures are available for human Toll-like receptors, which could represent appropriate modelling templates as essential residues are conserved in both metazoan and plant TNL proteins (Dinesh-Kumar et al. 2000). The TIR structure is predicted to form a five-stranded parallel β-sheet surrounded by five α-helices (Xu et al. 2000).
The CNL class of R proteins obtained their name because some non-TIR members contain predicted coiled-coil (CC) motifs, consisting of an α-helix-rich domain containing seven residue repeat sequences at their N-terminus (Pan et al. 2000). However, for the CC domain no information about its structure is available, and it is not clear how it folds or even whether it truly represents a coiled coil structure.
A recently recognised domain in the N-terminus of some NB-LRR proteins is the BED-finger (Aravind 2000; Tuskan et al. 2006). This domain is characterized by two motifs; one consisting of a pattern of cysteines and histidines that together might form a metal-chelating zinc finger, and the other containing a conserved tryptophane (Aravind 2000). Besides the name-giving Drosophila BEAF and DREF proteins that function as transcriptional regulators and chromatin insulators, this domain has been found in a subset of plant NB-LRR proteins and in DNA-binding proteins from tomato and tobacco (Aravind 2000). The presence of this BED-finger in DNA-binding domains and the prediction that it forms a zinc finger make it plausible that this structure represents a true DNA-binding domain. The observation of a DNA-binding domain in multi-domain STAND proteins (Leipe et al. 2004) perfectly fits their involvement in signal transduction and transcriptional regulation (see below).
The NB-ARC domain
As the ‘N’ in NB-ARC indicates, this domain has been predicted to bind nucleotides. This property is based on the presence of several conserved motifs characteristic for P-loop ATPases. Based on these motifs NB-LRRs, and many other proteins, could be classified as signal transduction ATPases with numerous domains (STAND) proteins (Leipe et al. 2004). The STAND protein family consists of five clades, the NB-ARC and NACHT proteins representing two of them (Leipe et al. 2004). All STAND proteins are multi-domain molecules that can contain DNA- or protein-binding domains, and super-repeat structures by which adaptor, regulatory switch, scaffolding, and, in some cases, signal-generating moieties are combined in a single protein. It was predicted that the STAND ATPase domain transmits conformational changes, induced by nucleotide exchange or hydrolysis, to the other domains of the protein thereby allowing it to generate a signal (Leipe et al. 2004). Biochemical studies on I-2, Mi-1.2 and N have indeed confirmed that the NB-ARC of these R proteins is a functional ATPase domain (Tameling et al. 2002; Ueda et al. 2006). The hydrolysis of ATP is likely to be accompanied by a conformational change of the NB-ARC domain, as after ATP-hydrolysis ADP-binding affinity increased dramatically, and because accumulation of mutant I-2 proteins in the ATP-bound state is likely to cause their autoactive phenotype (Tameling et al. 2006). These results are consistent with those obtained with the human NB-ARC protein Apaf-1, for which the various nucleotide binding states (either ADP or ATP) also represent different conformations. Cytochrome c binding to Apaf-1 triggers hydrolysis of the bound dATP and exchange of the formed dADP by dATP subsequently results in formation of the apoptosome that is able to trigger downstream signalling. Low (d)ATP levels result in the inability to exchange dADP and result in the formation of an inactive dADP-bound aggregate (Kim et al. 2005). The crystal structure of Apaf-1 revealed that the NB-ARC domain actually consists of four clearly distinguishable sub-domains (Riedl et al. 2005). These are the core P-loop NTPase fold, forming a five-stranded β-sheet flanked by α-helices, the ARC-1 domain, forming a four-helix bundle, the ARC-2 subdomains forming a winged-helix domain, and the ARC-3 subdomains, also forming a helical bundle. Specific ADP-binding is achieved through eight direct, and four H2O-mediated, interactions with various conserved residues present in the NB, ARC-1 and ARC-2 subdomains. These three subdomains are also conserved in R proteins, whereas the ARC-3 is lacking there (Albrecht and Takken 2006; Takken et al. 2006). As most of the residues involved in the interaction with the nucleotide as well as several peptide motifs are conserved in R proteins, this suggests a similar fold and possibly a similar molecular mechanism underlying their function. For an overview of the conserved motifs and domain structures we refer to recent reviews (McHale et al. 2006; Rairdan and Moffett 2007; Takken et al. 2006; van Ooijen et al. 2007).
Biochemical analysis of two auto-activating mutants of I-2, which induce plant defence responses in the absence of the pathogen, revealed that these mutants are affected in their ability to hydrolyse ATP, while the binding affinity for this nucleotide is not altered (Tameling et al. 2006). These data support a model in which there is a dynamic equilibrium between the ATP- and ADP-bound state of an NB-LRR R protein. In this model the ATP-bound state represents the active state and hydrolysis of the nucleotide flips the protein back to its inactive, ‘resting’ state (Takken et al. 2006; Tameling et al. 2006; van Ooijen et al. 2007). In this model at least two conformational changes of the protein are predicted to take place: exchange of ADP for ATP, resulting in the formation of the activated state, and subsequently hydrolysis of ATP whereby the protein returns to its resting state.
The first part of this model is analogous to that proposed for Apaf-1, in which exchange of bound dADP for dATP seems to be sufficient to allow apoptosome formation required for the initiation of apoptosis (Bao et al. 2007). However, it is not clear whether the proposed mechanism is generic for STAND proteins, as for instance ATPase activity has not been observed for the C. elegans analogue of APAF-1, CED-4, (Yan et al. 2005). Also for the plant TNL protein N, it has been suggested that not the exchange of ADP by ATP, but rather the hydrolysis of bound ATP is required for the protein to reach its active state (Ueda et al. 2006). Clearly there is a need for more biochemical data on the nucleotide-binding status of different and preferably full-length NB-LRR proteins to further explore the function of the NB-ARC domain. Although the structures are conserved, there will be differences in the underlying molecular mechanisms by which the various STAND proteins perform their function as molecular switches.
The carboxy-terminal extensions of NB-LRRs
Size and composition of the carboxy-terminal extensions differ between TNLs and CNLs. The latter often have short extensions of 40–80 amino acid residues, whereas the TNLs can have extensions of up to 300 amino acid residues (Meyers et al. 2003). In some cases these longer extensions have similarity to other proteins, the one example being Arabidopsis RRS1-R containing a WRKY domain and a nuclear localisation signal (NLS) at its carboxy terminus (Deslandes et al. 2003). A WRKY domain is also found in zinc-finger transcription factors and its name is derived from the conserved W-R-K-Y amino acid motif. For the majority of NB-LRRs, however, no recognizable domains have been observed in their C-terminal extensions and there are no known interactors of this domain.
Intramolecular interactions in NB-LRR R proteins
Activation of ETI, which is often accompanied by a cell death response, is costly for a plant and its proliferation could be fatal. Therefore this type of immunity has to be tightly regulated. One way of keeping NB-LRR proteins in check is by auto-inhibition, which seems to be accomplished by intramolecular interactions between the various domains. Deletion of the LRR domain of some NB-LRR proteins results in a weak auto-activation phenotype, indicative for a negative regulatory role of this domain (Bendahmane et al. 2002; Michael Weaver et al. 2006; Zhang et al. 2004). However, the LRRs can clearly also have a positive regulatory role, as expression of the N-terminal half of a CNL containing auto-activation mutations in the NB domain does not result in the activation of ETI unless the LRR is co-expressed (Moffett et al. 2002; Rairdan and Moffett 2006). Auto-activation mutants can not only be obtained by introducing specific point mutations in R proteins, but also by domain swaps between closely related paralogues as has been shown for Mi-1.2, Rx, Rp1 and L6 (Howles et al. 2005; Hwang et al. 2000; Rairdan and Moffett 2006; Sun et al. 2001). As these chimeras are combinations of wild-type domains, the observed auto-activation phenotype is likely to be due to incompatibility between regulatory subunits. These observations together support a model in which NB-LRR proteins are held in an auto-inhibited state by many (weak) interactions scattered over the various domains. Disturbance or misalignment of these interactions will release the auto-inhibition and allow the protein to proceed to its activated state. Evidence for such intramolecular interactions is provided by the observed association between the CC and NB-ARC-LRR domains of Rx, and between the LRR and the CC-NB-ARC domains in Bs-2 and Rx (Leister et al. 2005; Moffett et al. 2002; Rairdan and Moffett 2006). The first of the above interactions is dependent on a functional NB domain, supporting the model in which nucleotide exchange is required to release the signalling potential of the N-terminus. The latter interaction does not require a functional NB domain and appears to be mediated mainly by the ARC-1 sub-domain (Rairdan and Moffett 2006). One model for the activation of CNLs like Rx is based on the observed interaction between the LRR and the NB-ARC domain. Upon direct/indirect effector recognition, the interaction interface between the LRR and ARC-2 changes, allowing nucleotide exchange by the NB-ARC domain. This nucleotide exchange results in a conformational change of the NB-ARC and the N-terminal domain (Rairdan and Moffett 2007; Rairdan and Moffett 2006) thereby providing the means to convert recognition into signalling. How the activated NB-LRR protein subsequently activates defence signalling will be discussed below.
NB-LRRs and their putative function as transcriptional co-regulators in the nucleus
For a long time it has been thought that plant NB-LRR proteins would localise solely to the cytoplasm as no obvious nuclear localisation signal (NLS) was identified in these proteins. The finding that the atypical NB-LRR protein RRS1-R was present in the nucleus upon co-expression with its cognate effector potein PopP2, from Ralstonia solanacearum, was remarkable, but not totally surprising as this protein has a WRKY DNA binding domain and contains a predicted NLS. The two proteins interact in a yeast two-hybrid assay and co-localized exclusively in the nucleus when co-expressed. Co-expression with a PopP2 deletion mutant that lacked its bipartite NLS resulted in cytoplasmic localisation of both proteins. It is not yet clear whether the predicted NLS in RRS1-R is functional and required for nuclear localisation of RRS1-R (Deslandes et al. 2003; see below).
Two recent papers show that surprisingly also typical NB-LRR R proteins (tobacco TNL protein N and barley CNL protein MLA10) localise to the nuclear compartment (in addition to the cytoplasm) and that this localisation is required for activation of ETI (Burch-Smith et al. 2007; Shen et al. 2007). The potato CNL Rx also localises to both the cytoplasm and the nucleus, although it is currently unknown whether this localisation is required for Rx-mediated extreme resistance to potato virus X (PVX) (J. Bakker, personal communication). MLA10 confers resistance to Blumeria graminis f. sp. hordeii (Bgh) races that express the AvrA10 effector. Expression of AvrA10 in plant cells induces a physical association of MLA10 with WRKY transcription factor HvWRKY2 in the nucleus. In an earlier yeast two-hybrid screen HvWRKY1 and HvWRKY2 were identified as interactors of the CC domain of MLA10 and other MLA proteins containing identical N-termini. Both interactors belong to the WRKY family of transcription factors that bind specific W-box elements present in the promoters of many pathogen-responsive genes (Ulker and Somssich 2004). Shen et al. (2007) showed that HvWRKY1 and HvWRKY2 act as suppressors of PTI, as silencing of these genes resulted in an increased resistance whereas overexpression resulted in hyper-susceptibility to virulent Bgh races. Analysis of a double knock-out of the most closely related Arabidopsis WRKY genes (Atwrky18/Atwrky40) also revealed a role for these WRKYs as suppressors of PTI. Based on these data, a model was proposed that explains why the transcriptional reprogramming of ETI differs only quantitatively and kinetically from that of PTI (Shen et al. 2007) (Fig. 2c), as was suggested earlier (Tao et al. 2003). The difference in amplitude of the resistance response could also explain why ETI, but not PTI, is often associated with cell death, as proposed by Jones and Dangl 2006. The model presented in Fig. 2a implies negative regulation by transcriptional repressor proteins (e.g. WRKY transcription factors) to dampen the PTI response in order to prevent cell death. As described above specific microbes apparently evolved effector proteins by which they were able to suppress PTI and cause disease (Fig. 2b). When these or other effectors are recognised by host R proteins, ETI is induced. One way to achieve this is by relieving the negative regulatory system of PTI, which will result in faster and higher expression of PTI-triggered genes (Fig. 2c). This direct interaction and manipulation of transcriptional regulators by nuclear NB-LRR proteins could be a generic function of these proteins.
PAMP and effector-triggered immunity (PTI and ETI, respectively). a PAMPs/MAMPs can be recognised by receptor-like proteins (RLPs) that subsequently induce defence gene expression through positive (green) and negative (yellow) regulatory transcription factors (TRFs) eventually leading to PTI. b Many pathogens interfere with PTI by the production of effector molecules resulting in a diminished defence response. c Some of those effectors can be recognised by corresponding resistance proteins from the NB-LRR, RLP or RLK family resulting in an amplified form of the defence response termed ETI. For some NB-LRR R proteins nuclear localization is needed to trigger ETI. In that case NB-LRR proteins might facilitate the transcriptional reprogramming leading to ETI by direct interaction with transcriptional regulators in the nucleus
In the example above, MLA10 inhibits transcriptional repressors (HvWRKY1/2) upon pathogen perception. However, stimulation of positive transcriptional regulators is also a possibility as illustrated by the N protein that confers resistance to tobacco mosaic virus by recognition of viral P50 (Burch-Smith et al. 2007). Using yeast two-hybrid assays an SPL-type transcription factor was found to interact with N and to be required for N-mediated ETI (Dinesh-Kumar, personal communication). This implies that N could trigger ETI through direct interaction with a positive regulatory transcription factor (Fig. 2c). The Arabidopsis CNL R proteins RPM1 and RPP5 were also found to interact with a transcriptional regulator, called AtTIP49a, of which the animal homologue interacts with the TATA-binding protein complex. Silencing of this gene enhanced the weak ETI triggered by RPP5 and RPP2, but not the strong ETI mediated by RPM1 (Holt et al. 2002). As silencing of AtTIP49a did not lead to enhanced resistance to virulent pathogens, this protein might be a negative regulator of ETI, but not PTI. Whether other NB-LRRs could also function as transcriptional co-regulators remains to be investigated, but the presence of DNA binding domains in some NB-LRR proteins such as a WRKY or BED-finger domain (Fig. 1) supports this idea. In this respect, plant NB-LRRs could function similarly to the mammalian NACHT-LRR protein CIITA that translocates into the nucleus to regulate the expression of major histocompatibility complex (MHC) class II genes that are important for antigen presentation (Ting et al. 2006).
Nuclear NB-LRRs and nucleo-cytoplasmic trafficking in PTI and ETI
As no clear NLS sequences have been detected in most NB-LRRs it is unknown how they are translocated into the nucleus. One possibility is that they carry complex NLS sequences that are not easily predicted and deviate strongly from the classical NLS or the bipartite NLS (Gorlich and Kutay 1999). Another mechanism for import could be binding to a co-factor with an NLS sequence, a mechanism termed ‘piggyback’. This might be the case for RRS1-R, MLA10 and N, as they all interact with proteins that contain a functional NLS: PopP2, HvWRKY2 and an SPL-type transcription factor, respectively (Deslandes et al. 2003; Shen et al. 2007). However, the atypical RRS1-R protein could also be directly imported through recognition of its own NLS. For Rx, a Ran GTPase-activating protein 2 (RanGAP2) could serve as carrier according to the ‘piggyback’ mechanism. This protein has recently been identified as an Rx-associated protein by two different research groups (Sacco et al. 2007; Tameling and Baulcombe 2007). RanGAPs are highly conserved in eukaryotes and regulate the activity of the small GTPase Ran that is required for nucleo-cytoplasmic trafficking (Merkle 2003; Rose and Meier 2001). In interphase cells, localisation to the nuclear envelope (NE) is a feature of RanGAPs in both mammals and plants, which in the latter is mediated by the plant-specific WPP domain of RanGAP (Pay et al. 2002; Rose and Meier 2001). This is also the domain responsible for the interaction with the CC domain of Rx (Tameling and Baulcombe 2007). Specific silencing of RanGAP2 in Nicotiana benthamiana plants transgenic for Rx resulted in a loss of Rx-mediated extreme resistance to PVX and in local and systemic spread of the virus. RanGAP2 silencing did not affect N-mediated resistance to TMV or Pto/Prf-mediated resistance to Pseudomonas syringae pv. tabaci carrying the AvrPto effector, indicating that this protein might be specifically required for Rx (Tameling and Baulcombe 2007). Whether RanGAP2 indeed serves as a carrier or perhaps stimulates Rx import by recruiting yet another protein that serves as a carrier, remains to be investigated. An alternative hypothesis is that Rx activation modulates RanGAP2 activity in order to increase the nucleo-cytoplasmic trafficking of resistance co-factors involved in the induction of ETI.
Transport between the cytoplasm and the nucleus occurs exclusively through the nuclear pore complexes (NPCs) that are inserted in the nuclear envelope (Meier 2007; Merkle 2003). The NPCs are formed by large protein complexes containing nucleoporins. The precise composition of plant NPCs is not known, as most homologues of animal and yeast nucleoporins have not been identified in plants. Proteins of up to 40 kDa in molecular weight are able to diffuse through the NPCs, albeit much slower than the active transport mediated by the import and export receptors. NLS sequences are recognised by the import receptor importin (Imp) α, a member of the karyopherin family. Several karyopherin proteins have been identified in plants (Meier 2007). Docking of the importin cargo complex to the NE is mediated by another karyopherin family member, Imp β. Recently, in a mutagenesis screen, AtImpα3 has been shown to be required for the constitutive ETI mediated by suppressor of npr1–1 constitutive 1 (snc1), a TNL mutant that carries an auto-activating mutation (Palma et al. 2005). AtImpα3 is also required in wild-type Arabidopsis for PTI against virulent pathogens. A mutation in a second gene that was implicated in nucleo-cytoplasmic trafficking and required for snc1-mediated defence was identified and encodes an Arabidopsis nucleoporin 96 homolog that is important for both PTI and ETI (Zhang et al. 2004). These results indicate that nucleo-cytoplasmic trafficking plays an important role in both PTI and ETI, although further research is needed to identify which resistance co-factors are inhibited in their translocation to the nucleus when AtImpα3 and nucleoporin 96 function is abolished. It will be interesting to determine whether the tested NB-LRRs indeed localise to the nucleus and whether translocation of these might be inhibited in the described mutants. Although we are just starting to explore the role of nucleo-cytoplasmic trafficking in plant defence, it might turn out to play a crucial role in this process.
Crosstalk between R proteins classes
As mentioned above, some NB-LRR R proteins may initiate ETI by functioning as transcriptional co-regulators. How extracellular R proteins belonging to the RLP and RLK families (Fig. 1) could activate ETI is unclear. The founding members of the RLP class of R protein are the Cf proteins (Rivas and Thomas 2005). Due to their homology to the Arabidopsis RLP CLAVATA2 (CLV2) it was proposed that Cf proteins might relay downstream signalling by a similar mechanism (Joosten and De Wit 1999). CLV proteins control meristem development and function in a complex consisting of an RLP (CLV2), an RLK (CLV1) and an extracellular ligand (CLV3; Doerner 2003). Analogously, Cf proteins might depend on a plasma membrane-localised RLK for downstream signalling, with which it would form a heterodimer upon (indirect) perception of an extracellular effector protein of C. fulvum (indicated by a question mark in Fig. 2c). Although initial attempts to identify such Cf-protein complexes suggested the existence of ±400 kDA complexes for Cf-4 and Cf-9 in size exclusion chromatography experiments (Rivas et al. 2002a, b), later experiments revealed that the fast migration in the column is an intrinsic property of the Cf proteins (Van Der Hoorn et al. 2003). Therefore, it is currently unclear whether R proteins from either RLP or RLK class form heterodimers similar to the CLAVATA proteins.
An alternative approach to elucidate how RLP R proteins trigger downstream resistance signalling is via the identification of putative signal transduction components that are transcriptionally regulated upon the activation of Cf-mediated ETI. Gabriëls and associates performed a transcriptional profiling of tomato seedlings mounting Cf-4-mediated ETI (Gabriëls et al. 2006). A subset of these differentially expressed genes were silenced in transgenic N. benthamiana plants expressing Cf-4 to investigate their function in Cf-4-mediated HR. Interestingly this screen identified a gene coding for a CNL (Gabriëls et al. 2006). Since this protein was shown to be required for the Cf-4-mediated HR it was named NRC1 for NB-LRR protein required for HR-associated cell death 1 (see Fig. 2c, dotted arrow). NRC1 was not only required for RLP R proteins, but also for LeEIX (Gabriëls et al. 2007), an RLP that mediates recognition of ethylene-inducing xylanase (EIX), a potent elicitor of plant defence (Ron and Avni 2004). The genetic dependence of both RLP and RLK proteins on the same CNL suggests that they can trigger plant defences via a similar signalling pathway. It will be interesting to identify the cellular localisation of NRC1, since that would provide a clue on how RLP and RLKs affect defence gene expression.
Interestingly, NRC1 was also shown to be required for HR mediated by the CNL R proteins Prf, Rx and Mi-1.2 suggesting that also the signalling pathways of NB-LRR proteins are interwoven (Gabriëls et al. 2007). Additional support for such cross-talk is provided by the discovery of NRG1 (N-requirement gene 1), a CNL that is specifically required for the function of the TNL R protein N (Peart et al. 2005). Future studies should reveal whether crosstalk between CNLs, TNLs, RLPs and RLKs is a general phenomenon. If so, it could explain why the responses induced by the various R proteins are largely overlapping and depend on a limited number of conserved downstream signalling components (Martin et al. 2003; Tao et al. 2003).
Another major challenge for future studies is to solve the 3D structure of R proteins and to visualise their dynamics both at the subcellular as well as at the conformational level. A major bottleneck for biochemical and structural analyses of R proteins is the great difficulty to produce and purify sufficient amounts of intact and soluble native protein. A recent paper however, demonstrated the feasibility to use the methylotrophic yeast Pichia pastoris to produce and purify a relatively large amount of the almost full-length NB-LRR flax rust R protein M (Schmidt et al. 2007). If this protocol can also be applied for purification of other NB-LRR proteins it could provide the basis for experiments aimed to further our understanding of the mechanism by which these proteins operate in plant defence.
Abbreviations
- BED:
-
BEAF and DREF proteins zinc-finger DNA-binding domain
- CC:
-
Coiled Coil
- CNL:
-
CC-NB-LRR
- ETI:
-
effector-triggered immunity
- LRR:
-
leucine rich repeat
- MAMP:
-
microbe-associated molecular pattern
- NB-ARC:
-
nucleotide binding domain shared by Apaf-1, some R proteins and CED4
- PAMP:
-
pathogen-associated molecular pattern
- PTI:
-
PAMP-triggered immunity
- RLK:
-
receptor-like kinase
- RLP:
-
receptor-like protein
- SD:
-
solanaceous domain
- STAND:
-
signal transduction ATPases with numerous domains
- TIR:
-
Toll/interleukin-1 receptor like
- TNL:
-
TIR-NB-LRR
References
Ade, J., DeYoung, B. J., Golstein, C., & Innes, R. W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proceedings of the National Academy of Sciences of the United States of America, 104, 2531–2536.
Albrecht, M., & Takken, F. L. W. (2006). Update on the domain architectures of NLRs and R proteins. Biochemical and Biophysical Research Communications, 339, 459–462.
Aravind, L. (2000). The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends in Biochemical Sciences, 25, 421–423.
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved. Nauret Immunoogyl, 6, 973–979.
Axtell, M. J., Chisholm, S. T., Dahlbeck, D., & Staskawicz, B. J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Molecular Microbiology, 49, 1537–1546.
Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noel, L., Sadanandom, A., et al. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO Journal, 25, 2007–2016.
Bao, Q., Lu, W., Rabinowitz, J. D., & Shi, Y. (2007). Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Molecular Cell, 25, 181–192.
Bendahmane, A., Farnham, G., Moffett, P., & Baulcombe, D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site- leucine rich repeat protein encoded at the Rx locus of potato. Plant Journal, 32, 195–204.
Bieri, S., Mauch, S., Shen, Q. H., Peart, J., Devoto, A., Casais, C., et al. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495.
Birch, P. R., Rehmany, A. P., Pritchard, L., Kamoun, S., & Beynon, J. L. (2006). Trafficking arms: Oomycete effectors enter host plant cells. Trends in Microbiology, 14, 8–11.
Burch-Smith, T. M., Schiff, M., Caplan, J. L., Tsao, J., Czymmek, K., & Dinesh-Kumar, S. P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol, 5, e68.
Catanzariti, A. M., Dodds, P. N., & Ellis, J. G. (2007). Avirulence proteins from haustoria-forming pathogens. FEMS Microbiology Letters, 269, 181–188.
Chen, X., Shang, J., Chen, D., Lei, C., Zou, Y., Zhai, W., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant Journal, 46, 794–804.
Chisholm, S. T., Coaker, G., Day, B., & Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814.
De la Fuente van Bentem, S., Vossen, J. H., de Vries, K., van Wees, S. C., Tameling, W. I. L., Dekker, H., et al. (2005). Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant Journal, 43, 284–298.
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlothan, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proceedings of the National Academy of Sciences of the United States of America, 100, 8024-8029
Di Matteo, A., Federici, L., Mattei, B., Salvi, G., Johnson, K. A., Savino, C., et al. (2003). The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proceedings of the National Academy of Sciences of the United States of America, 100, 10124–10128.
Dinesh-Kumar, S. P., Tham, W. H., & Baker, B. J. (2000). Structure-function analysis of the tobacco mosaic virus resistance gene N. Proceedings of the National Academy of Sciences of the United States of America, 97, 14789–14794.
Doerner, P. (2003). Plant meristems: A merry-go-round of signals. Current Biology, 13, R368–R374.
Ellis, J. G., Dodds, P. N., & Lawrence, G. J. (2007). Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annual Review of Phytopathology, 45, 289–306.
Feys, B. J., & Parker, J. E. (2000). Interplay of signaling pathways in plant disease resistance. Trends in Genetics, 16, 449–455.
Gabriëls, S. H. E. J., Takken, F. L. W., Vossen, J. H., de Jong, C. F., Liu, Q., Turk, S. C., et al. (2006). CDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Molecular Plant-Microbe Interactions, 19, 567–576.
Gabriëls, S. H. E. J., Vossen, J. H., Ekengren, S. K., van Ooijen, G., Abd-El-Haliem, A. M., van den Berg, G. C. M., et al. (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. The Plant Journal, 50, 14–28.
Gorlich, D., & Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annual Review of Cell and Developmental Biology, 15, 607–660.
Grant, S. R., Fisher, E. J., Chang, J. H., Mole, B. M., & Dangl, J. L. (2006). Subterfuge and Manipulation: Type III Effector Proteins of Phytopathogenic Bacteria. Annual Review of Microbiology, 60, 425–449.
Holt 3rd, B. F., Belkhadir, Y., & Dangl, J. L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932.
Holt 3rd, B. F., Boyes, D. C., Ellerstrom, M., Siefers, N., Wiig, A., Kauffman, S., et al. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Developmental Cell, 2, 807–817.
Howles, P., Lawrence, G., Finnegan, J., McFadden, H., Ayliffe, M., Dodds, P., et al. (2005). Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Molecular Plant-Microbe Interactions, 18, 570–582.
Hubert, D. A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., et al. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO Journal, 22, 5679–5689.
Hwang, C. F., Bhakta, A. V., Truesdell, G. M., Pudlo, W. M., & Williamson, V. M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell, 12, 1319–1329.
Jones, J. D. G., & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329.
Joosten, M. H. A. J., & De Wit, P. J. G. M. (1999). The tomato-Cladosporium fulvum interaction: A versatile experimental system to study plant-pathogen interactions. Annual Review of Phytopathology, 37, 355–367.
Kajava, A. V. (1998). Structural diversity of leucine-rich repeat proteins. Journal of Molecular Biology, 277, 519–527.
Kim, H. E., Du, F., Fang, M., & Wang, X. (2005). Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proceedings of the National Academy of Sciences of the United States of America, 102, 17545–17550.
Kobe, B., & Deisenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends in Biochemical Sciences, 19, 415–421.
Leipe, D. D., Koonin, E. V., & Aravind, L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. Journal of Molecular Biology, 343, 1–28.
Leister, R. T., Dahlbeck, D., Day, B., Li, Y., Chesnokova, O., & Staskawicz, B. J. (2005). Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell, 17, 1268–1278.
Liu, Y., Burch-Smith, T., Schiff, M., Feng, S., & Dinesh-Kumar, S. P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. Journal of Biological Chemistry, 279, 2101–2108.
Liu, Y., Schiff, M., Serino, G., Deng, X. W., & Dinesh-Kumar, S. P. (2002). Role of SCF Ubiquitin-Ligase and the COP9 Signalosome in the N Gene- Mediated Resistance Response to Tobacco mosaic virus. Plant Cell, 14, 1483–1496.
Mackey, D., Holt, B. F., Wiig, A., & Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell, 108, 743–754.
Martin, G. B., Bogdanove, A. J., & Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annual Review of Plant Biology, 54, 23–61.
McHale, L., Tan, X., Koehl, P., & Michelmore, R. W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biology, 7, 212.
Meier, I. (2007). Composition of the plant nuclear envelope: Theme and variations. Journal of Experimental Botany, 58, 27–34.
Merkle, T. (2003). Nucleo-cytoplasmic partitioning of proteins in plants: Implications for the regulation of environmental and developmental signalling. Current Genetics, 44, 231–260.
Mestre, P., & Baulcombe, D. C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell, 18, 491–501.
Meyers, B. C., Dickerman, A. W., Michelmore, R. W., Sivaramakrishnan, S., Sobral, B. W., & Young, N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant Journal, 20, 317–332.
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., & Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell, 15, 809–834.
Moffett, P., Farnham, G., Peart, J., & Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO Journal, 21, 4511–4519.
Monosi, B., Wisser, R. J., Pennill, L., & Hulbert, S. H. (2004). Full-genome analysis of resistance gene homologues in rice. Theoretical & Applied Genetics, 109, 1434–1447.
Mucyn, T. S., Clemente, A., Andriotis, V. M. E., Balmuth, A. L., MucynOldroyd, G. E. D., Staskawicz, B. J., et al. (2006). The tomato NBARC-LRR protein Prf Interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell, 18 (10), 2792–806.
Nürnberger, T., & Kemmerling, B. (2006). Receptor protein kinases–pattern recognition receptors in plant immunity. Trends in Plant Scencei, 11, 519–522.
Palma, K., Zhang, Y., & Li, X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Current Biology, 15, 1129–1135.
Pan, Q., Wendel, J., & Fluhr, R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. Journal of Molecular Evolution, 50, 203–213.
Pay, A., Resch, K., Frohnmeyer, H., Fejes, E., Nagy, F., & Nick, P. (2002). Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant Journal, 30, 699–709.
Peart, J. R., Mestre, P., Lu, R., Malcuit, I., & Baulcombe, D. C. (2005). NRG1, a CC-NB-LRR Protein, together with N, a TIR-NB-LRR Protein, Mediates Resistance against Tobacco Mosaic Virus. Current Biology, 15, 968–973.
Rairdan, G. J., & Moffett, P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell, 18, 2082–2093.
Rairdan, G., & Moffett, P. (2007). Brothers in arms? Common and contrasting themes in pathogen perception by plant NB-LRR and animal NACHT-LRR proteins. Microbes and Infection, 9, 677–686.
Riedl, S. J., Li, W., Chao, Y., Schwarzenbacher, R., & Shi, Y. (2005). Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature, 434, 926–933.
Rivas, S., Mucyn, T., van den Burg, H. A., Vervoort, J., & Jones, J. D. G. (2002a). An approximately 400 kDa membrane-associated complex that contains one molecule of the resistance protein Cf-4. Plant Journal, 29, 783–796.
Rivas, S., Romeis, T., & Jones, J. D. G. (2002b). The Cf-9 disease resistance protein is present in an approximately 420- kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell, 14, 689–702.
Rivas, S., & Thomas, C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annual Review of Phytopathology, 43, 395–436.
Ron, M., & Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell, 16, 1604–1615.
Rose, A., & Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proceedings of the National Academy of Sciences of the United States of America, 98, 15377–15382.
Sacco, M., Mansoor, S., & Moffett, P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx and is required for Rx-mediated viral resistance. Plant Journal, DOI 10.111/j.1365-313x.2007.03213.x.
Schmidt, S. A., Williams, S. J., Wang, C. I., Sornaraj, P., James, B., Kobe, B., et al. (2007). Purification of the M flax-rust resistance protein expressed in Pichia pastoris. Plant Journal, 50, 1107–1117.
Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J. E., & Innes, R. W. (2003). Cleavage of Arabidopsis. PBS1 by a bacterial type III effector. Science, 301, 1230–1233.
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science, 315, 1098–1103.
Sun, X., Cao, Y., & Wang, S. (2006). Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiology, 140, 998–1008.
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae. in rice, encodes an LRR receptor kinase-like protein. Plant Journal, 37, 517–527.
Sun, Q., Collins, N. C., Ayliffe, M., Smith, S. M., Drake, J., Pryor, T., et al. (2001). Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics, 158, 423–438.
Takahashi, A., Casais, C., Ichimura, K., & Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 100, 11777–11782.
Takken, F. L. W., Albrecht, M., & Tameling, W. I. L. (2006). Resistance proteins: Molecular switches of plant defence. Current Opinion in Plant Biology, 9, 383–390.
Tameling, W. I. L., & Baulcombe, D. C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to potato virus X. Plant Cell, 19, 1682–694.
Tameling, W. I. L., Elzinga, S. D., Darmin, P. S., Vossen, J. H., Takken, F. L. W., Haring, M. A., et al. (2002). The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell, 14, 2929–2939.
Tameling, W. I. L., Vossen, J. H., Albrecht, M., Lengauer, T., Berden, J. A., Haring, M. A., et al. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiology, 140, 1233–1245.
Tan, X., Calderon-Villalobos, L. I., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature, 446, 640–645.
Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H. S., Han, B., et al. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell, 15, 317–330.
Ting, J. P., Kastner, D. L., & Hoffman, H. M. (2006). CATERPILLERs, pyrin and hereditary immunological disorders. Natures Review. Immunology, 6, 183–195.
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313, 1596–1604.
Ueda, H., Yamaguchi, Y., & Sano, H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Molecular Biology, 61, 31–45.
Ulker, B., & Somssich, I. E. (2004). WRKY transcription factors: From DNA binding towards biological function. Current Opinion in Plant Biology, 7, 491–498.
van der Biezen, E. A., & Jones, J. D. G. (1998). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Current Biology, 8, R226–227.
Van Der Hoorn, R. A., Rivas, S., Wulff, B. B., Jones, J. D., & Joosten, M. H. (2003). Rapid migration in gel filtration of the Cf-4 and Cf-9 resistance proteins is an intrinsic property of Cf proteins and not because of their association with high-molecular-weight proteins. Plant Journal, 35, 305–315.
van Ooijen, G., van den Burg, H. A., Cornelissen, B. J. C., & Takken, F. L. W. (2007). Structure and function of Resistance proteins in solanaceous plants. Annual Review of Phytopathology, 45, 43–72.
Weaver, M. L., Swiderski, M. R., Li, Y., & Jones, J. D. (2006). The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant Journal, 47, 829–840.
Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J. L., et al. (2000). Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature, 408, 111–115.
Yan, N., Chai, J., Lee, E. S., Gu, L., Liu, Q., He, J., et al. (2005). Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature, 437, 831–837.
Zhang, Y., Dorey, S., Swiderski, M., & Jones, J. D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant Journal, 40, 213–224.
Acknowledgements
The authors would like to acknowledge Savithramma Dinesh-Kumar and Jaap Bakker for sharing data before publication. We are grateful to Matthieu Joosten, Gerben van Ooijen and Martijn Rep for providing a critical review of the manuscript and helpful comments. Wladimir Tameling is supported by the EU-funded Integrated Project Bioexploit.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tameling, W.I.L., Takken, F.L.W. Resistance proteins: scouts of the plant innate immune system. Eur J Plant Pathol 121, 243–255 (2008). https://doi.org/10.1007/s10658-007-9187-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-007-9187-8