Abstract
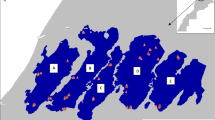
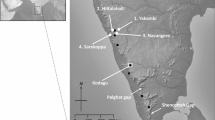
Forests are considered as one of the most complex terrestrial ecosystems due to their high level of biodiversity, including genetic diversity. Understanding the genetic diversity of keystone species at a population level is vital to forest managers and policymakers for the conservation and sustainable utilization of forest genetic resources. Quercus semecarpifolia, commonly known as brown oak, is a keystone species of climax community thriving in the alpine zone of the Himalayas, which is presently experiencing population decline and range shift under the changing climate. In the present study, a landscape genetic approach was employed for deciphering the population genetic structure of Q. semecarpifolia in the western Himalayas using nuclear simple sequence repeat (SSR) markers. By analysing 718 individuals of 24 populations at 10 SSR loci, a high gene diversity (expected heterozygosity, He = 0.72; Allelic richness, Ar = 8.37) was recorded with a moderate genetic differentiation (FST = 0.16; P < 0.001). Genetic clustering and STRUCTURE analysis have displayed two major gene pools which appear to be primarily differentiated by the landscape and ecological constraints rather than the linear geographical distances. The hierarchical AMOVA analysis further supports the regional genetic divergence with a substantial proportion of genetic variation detected among the regions. Diversity maps generated by spatial interpolation elucidated the distribution pattern of genetic diversity and structure across the range, and aid in the demarcation of the diversity hotspots for conservation implications. To the best of our knowledge, this is the first comprehensive genetic study carried out in any Himalayan oaks, and the information generated herein is novel and of paramount importance in guiding conservation decisions.



Similar content being viewed by others
Data availability
This manuscript did not generate any genetic resource in the form of nucleic acid or protein sequences, SNPs, expression data, etc. which could be deposited in the databases. The SSR sequences used here for the genotyping work are already in the public domain and their references have been cited in the manuscript. Further, all the data generated herein are presented either in the manuscript or supplementary material.
References
Allendorf FW, Luikart GH, Aitken SN (2013) Conservation and the genetics of populations, 2nd edn. Wiley-Blackwell
Bagnoli F, Vendramin GG, Buonamici A et al (2009) Is Cupressus sempervirens native in Italy? An answer from genetic and palaeobotanical data. Mol Ecol 18:2276–2286. https://doi.org/10.1111/j.1365294X.2009.04182.x
Bisht H (2001) Physiobiochemical aspects of seed viability in Quercus semecarpifolia smith: a possible recalcitrant seed. Ph D Thesis 45–48. https://doi.org/10.3923/rjss.2012.63.70. H.N.B. Garhwal University, Srinagar (Garhwal), 246174, Uttarakhand, India
Bisht VK, Kuniyal CP, Nautiyal BP, Prasad P (2013) Spatial distribution and regeneration of Quercus semecarpifolia and Quercus floribunda in a subalpine forest of western Himalaya, India. Physiol Mol Biol plants 19:443–448. https://doi.org/10.1007/s12298-013-0189-z
CBD (2022) Nations Adopt Four Goals, 23 Targets for 2030 In Landmark UN Biodiversity Agreement. 15th Conference of Parties to the UN Convention on Biological Diversity, 19th December 2022, Montreal. https://www.cbd.int/article/cop15-cbd-press-release-final-19dec2022. Accessed 1 June 2023
Chakraborty A, Saha S, Sachdeva K, Joshi PK (2018) Vulnerability of forests in the himalayan region to climate change impacts and anthropogenic disturbances: a systematic review. Reg Environ Chang 18:1783–1799. https://doi.org/10.1007/s11027-010-9257-7
Chiocchini F, Mattioni C, Pollegioni P et al (2016) Mapping the genetic diversity of Castanea sativa: exploiting spatial analysis for biogeography and conservation studies. J Geogr Inf Syst 8:248. https://doi.org/10.4236/jgis.2016.82022
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographic ranges: the central–marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Frankham R (2010) Where are we in conservation genetics and where do we need to go? Conserv Genet 11:661–663. https://doi.org/10.1007/s10592-009-0010-2
Gapare WJ (2014) Merging applied gene conservation activities with advanced generation breeding initiatives: a case of study of Pinus radiata D. Don. New For 45:311–331. https://doi.org/10.1007/s11056-013-9398-0
Geburek T, Konrad H (2008) Why the conservation of forest genetic resources has not worked. Conserv Biol 22:267–274. https://doi.org/10.1111/j.1523-1739.2008.00900.x
Gómez A, Vendramin GG, González-Martínez SC, Alía R (2005) Genetic diversity and differentiation of two Mediterranean pines (Pinus halepensis Mill. And Pinus pinaster Ait.) Along a latitudinal cline using chloroplast microsatellite markers. Divers Distrib 11:257–263. https://doi.org/10.1111/j.1366-9516.2005.00152.x
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467. https://doi.org/10.1111/j.1461-0248.2005.00739.x
Hamrick JL, Godt MJW (1990) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer Associates Inc., Sunderland, pp 43–63
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc B 351:1291–1298. https://doi.org/10.1098/rstb.1996.0112
Hengl T (2009) A practical guide to geostatistical mapping. University of Amsterdam, Amsterdam
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Joshi PK, Rawat A, Narula S, Sinha V (2012) Assessing impact of climate change on forest cover type shifts in western himalayan eco-region. J For Res 23:75–80. https://doi.org/10.1007/s11676-012-0235-7
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kremer A, Hipp AL (2020) Oaks: an evolutionary success story. New Phytol 226:987–1011. https://doi.org/10.1111/nph.16274
Lee Y-J, Hwang S-Y, Ho K-C, Lin T-P (2006) Source populations of Quercus glauca in the last glacial age in Taiwan revealed by nuclear microsatellite markers. J Hered 97:261–269. https://doi.org/10.1093/jhered/esj030
Lemes MR, Gribel R, Proctor J, Grattapaglia D (2003) Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the brazilian Amazon, based on variation at microsatellite loci: implications for conservation. Mol Ecol 12:2875–2883. https://doi.org/10.1046/j.1365-294X.2003.01950.x
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760. https://doi.org/10.1046/j.1523-1739.1995.09040753.x
Li YL, Liu JX (2018) StructureSelector: a web-based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour 18:176–177. https://doi.org/10.1111/1755-0998.12719
López-Cortegano E, Pouso R, Labrador A et al (2019) Optimal management of genetic diversity in subdivided populations. Front Genet 10:843. https://doi.org/10.3389/fgene.2019.00843
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 65–95. https://doi.org/10.1146/annurev.es.15.110184.000433
Lupini A, Aci MM, Mauceri A et al (2019) Genetic diversity in old populations of sessile oak from Calabria assessed by nuclear and chloroplast SSR. J Mt Sci 16:1111–1120. https://doi.org/10.1007/s11629-018-5335-1
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Matschiner M, Salzburger W (2009) TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25:1982–1983. https://doi.org/10.1093/bioinformatics/btp303
McDermott JM, McDonald BA (1993) Gene Flow in Plant Pathosystems. Annu Rev Phytopathol 31:353–373. https://doi.org/10.1146/annurev.py.31.090193.002033
Milgroom MG (2015) Migration and population structure. In: Population Biology of Plant Pathogens: Genetics, Ecology, and Evolution. The American Phytopathological Society, pp 119–146. https://doi.org/10.1094/9780890544525.006
Mishima K, Watanabe A, Isoda K et al (2006) Isolation and characterization of microsatellite loci from Quercus mongolica var. Crispula. Mol Ecol Notes 6:695–697. https://doi.org/10.1111/j.1471-8286.2006.01313.x
Misiewicz TM, Fine PVA (2014) Evidence for ecological divergence across a mosaic of soil types in an amazonian tropical tree: Protium subserratum (Burseraceae). Mol Ecol 23:2543–2558. https://doi.org/10.1111/mec.12746
Morente-López J, García C, Lara-Romero C et al (2018) Geography and environment shape landscape genetics of Mediterranean alpine species Silene ciliata Poiret. (Caryophyllaceae). Front Plant Sci 1698. https://doi.org/10.3389/fpls.2018.01698
Negi SS, Naithani HB (1995) Oaks of India, Nepal and Bhutan. International Book Distributors, Dehradun
Negi M, Negi VS (2021) Temporal changes in Oak forests over last three decades in western Himalaya, India. Trees For People 6:100146. https://doi.org/10.1016/j.tfp.2021.100146
Negi VS, Pathak R, Rawal RS, Bhatt ID, Sharma S (2019) Long-term ecological monitoring on forest ecosystems in indian Himalayan Region, Criteria and indicator approach. Ecol Indic 102:374–381. https://doi.org/10.1016/j.ecolind.2019.02.035
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170. https://doi.org/10.1007/BF02300753
Nixon KC (2006) Global and neotropical distribution and diversity of oak (genus Quercus) and oak forests. In: Kappelle M (ed) Ecology and conservation of neotropical montane oak forests. Springer, Berlin, Heidelberg, pp 3–13
Ohsawa T, Tsuda Y, Saito Y et al (2007) Altitudinal genetic diversity and differentiation of Quercus crispula in the Chichibu Mountains, central Japan. Int J Plant Sci 168:333–340. https://doi.org/10.1086/510413
Oldenbroek JK (2017) Genomic management of animal genetic diversity. Wageningen Academic Publishers, Wageningen
Oyama K, Ramírez-Toro W, Peñaloza-Ramírez JM et al (2018) High genetic diversity and connectivity among populations of Quercus candicans, Quercus crassifolia, and Quercus castanea in a heterogeneous landscape in Mexico. Trop Conserv Sci 11. https://doi.org/10.1177/19400829187661
Pandey M, Rajora OP (2012) Genetic diversity and differentiation of core vs. peripheral populations of eastern white cedar, Thuja occidentalis (Cupressaceae). Am J Bot 99:690–699. https://www.jstor.org/stable/41415610
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537e2539. https://doi.org/10.1093/bioinformatics/bts460
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855. https://doi.org/10.1111/j.1523-1739.1998.96489.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://pubmed.ncbi.nlm.nih.gov/10835412/
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
Rawat B, Rawat JM, Purohit S et al (2022) A comprehensive review of Quercus semecarpifolia Sm.: an ecologically and commercially important himalayan tree. Front Ecol Evol 10:961345. https://doi.org/10.3389/fevo.2022.961345
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x
Reis TS, Ciampi-Guillardi M, Bajay MM et al (2015) Elevation as a barrier: genetic structure for an atlantic rain forest tree (Bathysa australis) in the Serra do marmountain range, SE Brazil. Ecol Evol 5:1919–1931. https://doi.org/10.1002/ece3.1501
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Saran S, Joshi R, Sharma S et al (2010) Geospatial modeling of Brown oak (Quercus semecarpifolia) habitats in the Kumaun Himalaya under climate change scenario. J Indian Soc Remote Sens 38:535–547. https://doi.org/10.1007/s12524-010-0038-2
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis. User Man ver 2:2496–2497. https://journals.sagepub.com/doi/pdf/10.1177/117693430500100003
Shekhar C, Rawat A, Bhandari MS et al (2021) Cross-transferability-based identification and validation of simple sequence repeat (SSR) markers in oaks of western Himalayas. Silvae Genet 70:108–116. https://doi.org/10.2478/sg-2021-0009
Shekhar C, Ginwal HS, Meena RK et al (2022) Spatio-temporal distribution of broad-leaved Quercus semecarpifolia indicates altitudinal shift in northwestern Himalayas. Plant Ecol 223:671–697. https://doi.org/10.1007/s11258-022-01240-x
Shepard D (1968) A two-dimensional interpolation function for irregularly-spaced data. In: Proceedings of the 1968 23rd ACM national conference. Association for Computing Machinery, New York, pp 517–524
Shrestha BB (2003) Quercus semecarpifolia Sm. In the himalayan region: Ecology, exploitation and threats. Himal J Sci 1:126–128. https://doi.org/10.3126/hjs.v1i2.212
Shrestha UB, Gautam S, Bawa KS (2012) Widespread climate change in the Himalayas and associated changes in local ecosystems. PLoS ONE 7:e36741. https://doi.org/10.1371/journal.pone.0036741
Singh SP (2018) Research on indian Himalayan Treeline Ecotone: an overview. Trop Ecol 59(2):163–176. http://www.tropecol.com/pdf/open/PDF_59_2/01%20Surender%20P%20Singh.pdf
Singh G, Rawat GS, Verma D (2010) Comparative study of fuelwood consumption by villagers and seasonal “Dhaba owners” in the tourist affected regions of Garhwal Himalaya, India. Energy Policy 38:1895–1899. http://www.ias.ac.in/currsci
Singh G, Rai ID, Rawat GS (2011) The year 2010 was’ mast seed year’for the Kharsu oak (Quercus semecarpifolia Sm.) In the western Himalaya. Curr Sci 100:1275–1276. http://www.ias.ac.in/currsci
Souto C, Mathiasen P, Acosta M (2015) Identifying genetic hotspots by mapping molecular diversity of widespread trees: when commonness matters. J Hered 106:537–545. https://doi.org/10.1093/jhered/esv023
Spence ES, Fant JB, Gailing O et al (2021) Comparing genetic diversity in three threatened Oaks. Forests 12:561. https://doi.org/10.3390/f12050561
Takezaki N, Nei M, Tamura K (2010) POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27:747–752. https://doi.org/10.1093/molbev/msp312
Tashi S (2004) Regeneration of Quercus semecarpifolia Sm. in an old growth oak forest under Gidakom FMU-Bhutan. M. Sc. Dissertation, Forest Ecology and Forest Management. Department of Forestry, Wageningen University and Research Centrum, the Netherlands
Templeton AR, Routman E, Phillips CA (1995) Separating population structure from population history: a cladistic analysis of the geographic distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140:767–782. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1206651/
Tewari A, Shah S, Singh N et al (2019) Acorn maturation and regeneration problem in Quercus semecarpifolia Sm. Himal tree line Int J Sci Technol Res 8:3781–3787
Ueno S, Tsumura Y (2008) Development of ten microsatellite markers for Quercus mongolica var. Crispula by database mining. Conserv Genet 9:1083–1085. https://doi.org/10.1007/s10592-007-9462-4
Ueno S, Taguchi Y, Tsumura Y (2008) Microsatellite markers derived from Quercus mongolica var. Crispula (Fagaceae) inner bark expressed sequence tags. Genes Genet Syst 83:179–187. https://doi.org/10.1266/ggs.83.179
Uttarakhand at a Glance 2020-21 (2021) Directorate of Economics &Statistics, Government of Uttarakhand, Dehradun, India. 31 pages. https://des.uk.gov.in/pages/display/61-uttarakhnad-at-a-glance
Uttarakhand Forest Statistics 2014–2015 (2015) Uttarakhand Forest Department, Dehradun, India. 181 pages. http://www.uafdc.in/Acts&Rules/Forest_Statistical_2014-15.pdf
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Vetaas OR (2000) The effect of environmental factors on the regeneration of Quercus semecarpifolia Sm. In central Himalaya, Nepal. Plant Ecol 146:137–144. https://doi.org/10.1023/A:1009860227886
Wright S (1943) Isolation by distance. Genetics 28:114–138. https://doi.org/10.1093/genetics/28.2.114
Wright S (1969) Evolution and the genetics of populations, volume 2: the theory of gene frequencies. University of Chicago Press, Chicago
Wright HE Jr (1976) The Environmental setting for Plant Domestication in the Near East: wild cereal grains may not have entered the Near East until the end of the Pleistocene 11,000 years ago. Sci (80-) 194:385–389. https://doi.org/10.1126/science.194.4263.385
Yang J, Vázquez L, Feng L et al (2018) Climatic and soil factors shape the demographical history and genetic diversity of a deciduous oak (Quercus liaotungensis) in Northern China. Front Plant Sci 9:1534. https://doi.org/10.3389/fpls.2018.01534
Acknowledgements
This research was funded by the Ministry of Environment Forests and Climate Change, Govt. of India through National CAMPA grant awarded to FRI Dehradun in the form of a research project. We would like to thank the Director, Forest Research Institute, Dehradun for providing all infrastructure and logistic support for conducting this work. Also, we would like to extend our gratitude to state forest departments of Uttarakhand for providing permissions and support during field surveys and sample collection. At last, we duly acknowledge the contribution of anonymous reviewers for imparting their knowledge and wisdom into this manuscript.
Funding
This research was funded by Ministry of Environment Forests and Climate Change, Govt. of India through National CAMPA grant awarded to FRI Dehradun in the form of a research project.
Author information
Authors and Affiliations
Contributions
HSG and RKM, conceptualized the problem, interpreted data and prepared final manuscript. CS, RS, AY, MSB, SB, RK and RKM performed the field sampling. AR and HK performed laboratory work and analyzed the marker data. MSB and RS carried out geo-spatial analysis. Further, all the authors contributed in preparation and editing of the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest/Competing interests
I hereby affirm that the manuscript has neither published elsewhere nor submitted simultaneously for publication elsewhere. The article has been read by each author and there is no conflict of interest.
Ethics approval
All necessary permissions were obtained for sample collection from concerned state forest department.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
: Spatial overlaying of private allelic richness (PAr) over distribution map of Q. semecarpifolia. The areas or populations depicted in dark red shows presence of high proportion of private alleles. Fig. S2: Relationship of pairwise genetic distance with the pairwise geographic distances: a and b) shows the relationship of genetic distance with horizontal and vertical distance when analyzed all the 24 populations combined, c and d) shows relationship of genetic distance with horizontal and vertical distance when analyzed the seven populations of Cluster I separately, e and f) shows relationship of genetic distance with horizontal and vertical distance when analyzed the remaining 17 populations of Cluster II separately. Fig S3: Graphical representation of the estimated Ln probability of data (a) and ΔK (b) in relation to the set K values from 1-10. Fig S4: Principal coordinate analysis (PCoA) showing sub-clustering of Q. semecarpifolia populations of Cluster 1 (a) and Cluster II (b). Fig S5: STRUCTURE analysis of the Q. semecarpifolia populations of Cluster I: a) Graphical representation of the ΔK in relation to the set K values from 1-10; b-c) Bar plot showing pattern of genetic admixture among individual genotypes and populations at K=2 (b) and K=5. Each population separated by a vertical line are labelled with a number below which correspond the code given in table 1. Fig S6: STRUCTURE analysis of the Q. semecarpifolia populations of Cluster II: a) Graphical representation of the ΔK in relation to the set K values from 1-10; b) Bar plot showing pattern of genetic admixture among individual genotypes and populations at K=3. Each population separated by a vertical line are labelled with a number below which correspond the code given in table 1. Table S1. Comparison of diversity indices calculated in this study for Quercus semecarpifolia (Sl. No. 1) and other oaks species studied earlier
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ginwal, H.S., Rawat, A., Shekhar, C. et al. Population genetic structure of a timberline oak (Quercus semecarpifolia Sm.) of western Himalayas and conservation implications. Conserv Genet 25, 133–147 (2024). https://doi.org/10.1007/s10592-023-01558-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01558-7