Abstract
The early and accurate diagnosis of brain tumors is critical for effective treatment planning, with Magnetic Resonance Imaging (MRI) serving as a key tool in the non-invasive examination of such conditions. Despite the advancements in Computer-Aided Diagnosis (CADx) systems powered by deep learning, the challenge of accurately classifying brain tumors from MRI scans persists due to the high variability of tumor appearances and the subtlety of early-stage manifestations. This work introduces a novel adaptation of the EfficientNetv2 architecture, enhanced with Global Attention Mechanism (GAM) and Efficient Channel Attention (ECA), aimed at overcoming these hurdles. This enhancement not only amplifies the model’s ability to focus on salient features within complex MRI images but also significantly improves the classification accuracy of brain tumors. Our approach distinguishes itself by meticulously integrating attention mechanisms that systematically enhance feature extraction, thereby achieving superior performance in detecting a broad spectrum of brain tumors. Demonstrated through extensive experiments on a large public dataset, our model achieves an exceptional high-test accuracy of 99.76%, setting a new benchmark in MRI-based brain tumor classification. Moreover, the incorporation of Grad-CAM visualization techniques sheds light on the model’s decision-making process, offering transparent and interpretable insights that are invaluable for clinical assessment. By addressing the limitations inherent in previous models, this study not only advances the field of medical imaging analysis but also highlights the pivotal role of attention mechanisms in enhancing the interpretability and accuracy of deep learning models for brain tumor diagnosis. This research sets the stage for advanced CADx systems, enhancing patient care and treatment outcomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
There are illnesses that pose a serious hazard to human life. Cancer is one of the significant illnesses among them. A condition known as cancer occurs when cells in a particular body part multiply and expand uncontrolled. It is estimated that in 2024, there will be approximately 2,001,140 new cancer cases and 611,720 deaths related to cancer in the United States [1]. Various types of cancer exist, including brain tumors, which are categorized as malignant due to their malignant growth in the brain [2]. This differs from benign brain tumors, which are non-malignant and typically grow at a slower pace [3]. The growth rate, type, stage of advancement, and location of a brain tumor are among the characteristics that determine whether it is malignant or benign [4, 5]. It is expected that 700,000 Americans already have primary brain tumors, and that 94,390 more Americans will be diagnosed with main brain tumors in 2023 [6].
Tumors in the human brain are found using a variety of diagnostic techniques, both invasive and non-invasive [2, 3]. By way of illustration, a biopsy is an intrusive procedure that entails extracting a sample through a surgical cut and studying it under a microscope to identify malignancy (malignant tumor). Brain tumor biopsies, on the other hand, are normally not carried out prior to definitive brain surgery, in contrast to malignancies in other sections of the body [3]. In order to diagnose brain tumors, non-invasive imaging methods like magnetic resonance imaging (MRI), positron emission tomography, and computed tomography are thought to be quicker and safer. The capacity of MRI to offer comprehensive information regarding the location, development, shape, and size of the brain tumor in both 2D and 3D formats with high resolution makes it the method of choice among all the non-invasive imaging modalities discussed [7, 8]. Due to the high patient volume, manual interpretation of MRI images is time-consuming for medical professionals and prone to mistakes. According to Fig. 1, an MRI image frequently used to represent various medical conditions (A) displays a suspicious irregularity in the lower right corner. (B, C) on the other hand, show a malignant situation, with the tumor taking up more space. In contrast to earlier stages, (D) exhibits tumor growth and the eradication of nearby cells [9].
To combat cancer, it is crucial to utilize early detection methods for both early-stage detection of the disease and the implementation of various preventive measures. One of the most popular techniques for early tumor identification in this context is the MRI technology [8]. Based on their experience, radiologists routinely use this technique to detect brain cancers early. There is a clear need for new methods that can expedite and enhance decision-making processes in the field of medical imaging [10]. The medical field extensively utilizes new technologies, with artificial intelligence and machine learning gaining increasing preference due to their ability to perform rapid processing and achieve high accuracy rates in disease diagnosis [11, 12].
Artificial intelligence is a revolutionary technology that has a profound impact on our lives [13, 14]. The automation of image processing can greatly benefit from approaches based on machine learning and innovative computational methods. A subfield of artificial intelligence, deep learning is commonly applied on large datasets to automatically extract features, thus gaining considerable popularity for its ability to perceive complex patterns and relationships within data [15]. These algorithms can autonomously tackle numerous complex tasks in fields such as medical image processing, drug discovery, agriculture, and defense, achieving high levels of success [16,17,18,19,20]. Deep learning-supported techniques are beneficial, especially in key fields like biological image analysis, where processing speed is crucial, and the cost and hazards of misinterpretation are significant [19]. To increase productivity and the accuracy of diagnoses, computer-aided approaches are replacing traditional medical image analysis techniques. Due to the well-known efficacy of deep learning-based computer-aided diagnosis solutions, deep learning-based medical image analysis is a significant and active research area, with many researchers working in this field [21].
In CAD applications, deep learning algorithms, notably Convolutional Neural Networks (CNNs), represent a significant leap forward compared to traditional machine learning methods [22]. These conventional techniques often rely on manually crafted features, a process that is not only time-consuming but also heavily dependent on the expertise of domain specialists. CNNs, however, revolutionize this landscape by autonomously extracting relevant features from images, effectively bypassing the need for labor-intensive feature engineering. The superiority of CNNs is particularly evident in medical image analysis, where they have garnered widespread acclaim for their remarkable performance [23, 24]. Specifically, CNNs have emerged as powerful tools for accurately identifying various types of brain tumors and other medical conditions. What sets CNNs apart is their innate ability to discern critical information directly from images, thereby obviating the need for human intervention in feature selection [25]. This automated feature extraction capability not only streamlines the CAD process but also enhances its accuracy and efficiency [26,27,28]. By harnessing CNNs, CAD systems can swiftly and reliably categorize medical images with precision, thus facilitating quicker diagnoses and treatment decisions [29]. Moreover, CNNs empower healthcare professionals by providing them with insights derived directly from raw data, facilitating a more informed approach to patient care [30].
Moreover, vision transformers, which present a distinct architecture from CNNs, have exhibited promising outcomes across diverse domains, including diseases related to brain tumors [31]. Unlike CNNs, vision transformers utilize an attention mechanism to capture distant dependencies and relationships among image patches, thereby enabling them to adeptly model intricate visual patterns. This architectural approach has showcased remarkable efficiency in applications spanning natural language processing and has recently garnered attention in computer vision tasks. In the context of brain tumor classification, vision transformers have demonstrated their capability to capture both global and local image characteristics, facilitating a more comprehensive and precise analysis. This ability to integrate information from various spatial scales enhances their effectiveness in discerning subtle nuances within medical images, contributing to improved diagnostic accuracy and treatment planning. Consequently, vision transformers emerge as a promising alternative to CNNs, offering advanced capabilities for image analysis tasks in healthcare and beyond [32].
Numerous scientific studies focus on brain tumor diagnosis, revealing that deep learning has led to significant advancements in this area [9, 33,34,35,36,37]. Deep learning has become a groundbreaking method, offering crucial benefits for accurately diagnosing brain tumors due to their complexity. These models autonomously extract complex patterns and features from large medical datasets like MRI scans, enabling precise tumor segmentation and classification [33, 34, 38]. After examining various reviews and surveys, it’s clear that deep learning has led to numerous significant discoveries in brain tumor diagnosis. Research indicates that deep learning has evolved into an innovative and impactful approach in this field. Bhagyalaxmi et al. [36] found that deep learning, particularly CNNs, shows promise for brain tumor detection. While their review outlines current techniques, they suggest future research explore hybrid models combining deep learning with other methodologies to maximize effectiveness. Awuah et al. [39] highlights AI’s potential to enhance patient outcomes in neurosurgery by aiding neurosurgeons in diagnostics, prognostics, and surgical decision-making. AI, including machine learning and deep learning, has advanced significantly, reducing complications, improving surgical planning, and overall benefiting patient care in neurosurgery. Levy et al. [40] highlight the significant articles on machine learning in neurosurgery. They emphasize machine learning’s application across various sub-specialties to enhance patient care, particularly in outcome prediction, patient selection, and surgical decision-making.
Among the limitations of deep learning for brain tumor classification are the scarcity of labeled data, incomplete model optimization, the selection of appropriate models, overfitting, interpretability issues, and data imbalance between classes. These constraints can negatively impact the overall performance and accuracy of deep learning models in tumor classification. Specifically, overfitting may diminish the model’s generalization ability, while challenges in interpretability can complicate trust and acceptance in medical applications. Nonetheless, awareness of these limitations presents an opportunity for the development of more advanced methods and strategies to enhance brain tumor classification. Thus, deep learning models can be more effectively utilized in this domain.
In the field of medical image recognition and segmentation, particularly in MRI-based brain tumor detection, significant advancements have been achieved through deep learning technologies. Despite these advancements, current methods display considerable limitations in effectiveness and acceptance within clinical settings. These challenges include limited access to sufficient and diverse data for deep learning architectures, the frequent absence of a test set in data partitioning, the inability to precisely detect early-stage tumors, high variability in tumor appearances across patients, and the “black box” nature of many deep learning solutions. The latter poses a particularly significant challenge, as transparency and interpretability are crucial in clinical applications for building trust and transforming insights into actionable decisions.
The primary motivation of this research stems from the urgent need to improve the diagnosis of brain tumors, one of the most lethal cancer types, to enhance patient survival rates, as well as to overcome the aforementioned obstacles. Additionally, the necessity arises from the lack of detailed exploration into attention mechanisms for brain tumors. This study aims to introduce an innovative CNN architecture enhanced with attention mechanisms, presenting a critical alternative that surpasses the performance of both vision transformer and conventional CNN-based models. By partitioning datasets into training, validation, and test sets, this work seeks to significantly advance the detection of brain tumors through MRI scans. This approach is grounded in the importance of providing a more sensitive detection of early-stage tumor indicators, improving adaptability to the broad spectrum of tumor presentations among different patients, and enhancing model interpretability to support clinical decision-making processes. This study makes significant contributions to the field of deep learning for MRI-based brain tumor classification, notably advancing current methodologies in terms of precision, adaptability, and clinical applicability. The research is particularly distinguished by the following key contributions:
-
Integrating attention mechanisms within the EfficientNetv2 [41] framework, the study presents an advanced CNN architecture tailored specifically for the complexities of brain tumor imagery in MRI scans. The incorporation of the Global Attention Mechanism (GAM) [42] and Efficient Channel Attention (ECA) [43] represents an innovative approach to deep learning models.
-
A notable contribution of this work is its enhanced sensitivity towards early-stage tumors. The proposed model’s capability to detect subtle features indicative of early tumor development signifies a significant advancement over existing models that often struggle with such precision.
-
Addressing the challenge of variability in tumor appearance across patients, the model’s redesign for brain tumor diagnosis through the application of attention mechanisms enables it to adapt effectively to a wide range of tumor presentations, substantially increasing its generalization ability in classification.
-
By incorporating Gradient-weighted Class Activation Mapping (Grad-CAM) [44] visualization techniques, the study takes a new step in making deep learning models more interpretable. This contribution aids in validating and explaining the model’s decision-making process and enhances the reliability and trust in automated diagnoses within clinical settings.
-
Achieving a high-test accuracy of 99.76% on an extensive public dataset, the research sets a new precedent in MRI-based brain tumor classification.
-
The study compares the performance of a total of 45 deep learning models, including state-of-the-art technologies such as EfficientNet, MobileNetv3, InceptionNext, RepGhostNet, MobileViTv2, DeiT3, MaxViT, ResMLP, FastViT, and MetaFormer, based solely on test data.
-
Finally, it is the first study to apply the performance of the EfficientNet model and various attention mechanisms for the identification of brain tumors.
The subsequent sections of the paper are organized as follows: Sect. 2 conducts a literature review, examining studies on brain tumors utilizing deep learning techniques. In Sect. 3, a comprehensive description of the materials, datasets, and the proposed framework used in the approach is provided, along with a brief analysis of all models employed. Section 4 presents a thorough performance evaluation of the proposed approach, encompassing both quantitative and qualitative studies. This section emphasizes the significance of Grad-CAM methods in offering valuable insights into the decision-making process of artificial intelligence models. Additionally, Sect. 4 includes a comparative analysis of brain tumor classification models. Finally, Sect. 5 discusses the results obtained from the proposed approach.
2 Related works
Deep learning is a groundbreaking method for accurately diagnosing complex brain tumors. These models autonomously extract patterns from large MRI datasets, enabling precise tumor segmentation and classification. Deep learning’s ability to handle vast amounts of data swiftly and accurately enhances diagnostic effectiveness, speeds up treatment decisions, and improves patient outcomes. However, successful integration into clinical practice requires close collaboration between AI experts and medical professionals to ensure trust, interpretability, and the use of deep learning as a supportive tool rather than a replacement for medical expertise. In the field of brain tumors, numerous deep learning-based CADx methods have been developed. Some of these studies include the followings.
Ullah et al. [45] introduced the Multiscale Residual Attention-UNet (MRA-UNet) for brain tumor segmentation, utilizing three consecutive slices and multiscale learning. Postprocessing techniques enhanced segmentation accuracy, yielding state-of-the-art results on BraTS2017, BraTS2019, and BraTS2020 datasets with average dice scores of 90.18, 87.22, and 86.74% for whole tumor, tumor core, and enhanced tumor regions, respectively. Celik and Inik [31] proposed a hybrid method for brain tumor classification, combining a novel CNN model for feature extraction with ML algorithms for classification. They compared CNN performance using nine state-of-the-art models and optimized ML algorithm hyperparameters with Bayesian optimization. Results show the hybrid model achieves 97.15% mean classification accuracy, along with 97% recall, precision, and F1-score values. Anantharajan et al. [46] propose a novel method for MRI brain tumor detection using DL and ML. They preprocess MRI images with the Adaptive Contrast Enhancement Algorithm (ACEA) and median filter, then segment them with fuzzy c-means. Features like energy, mean, entropy, and contrast are extracted using the Gray-level co-occurrence matrix (GLCM). Abnormal tissues are classified using the Ensemble Deep Neural Support Vector Machine (EDN-SVM) classifier. Pacal [10] introduced an advanced deep learning method based on the Swin Transformer. It incorporates a Hybrid Shifted Windows Multi-Head Self-Attention module (HSW-MSA) and a rescaled model to enhance accuracy, reduce memory usage, and simplify training complexity. By replacing the traditional MLP with the Residual-based MLP (ResMLP), the Swin Transformer achieves improved accuracy, training speed, and parameter efficiency. Evaluation on a publicly available brain MRI dataset demonstrates enhanced performance through transfer learning and data augmentation techniques for efficient and robust training.
Remzan et al. [47] addressed brain tumor classification using deep learning algorithms, enhancing diagnostic precision in radiology. They proposed RadImageNet and employed transfer learning to overcome challenges with limited and non-diverse medical image datasets. Their ensemble learning strategies achieved notable accuracy improvements, reaching 97.71 and 97.40% accuracy with ResNet-50 and DenseNet121 features, respectively. Ullah et al. proposed MRA-UNet, a novel fully automated method for brain tumor segmentation. It utilizes multiscale learning and sequential information from three consecutive slices. MRA-UNet accurately segments enhanced and core tumor regions by employing an adaptive region of interest scheme. Postprocessing techniques like conditional random field and test time augmentation further enhance its performance [45]. Based on an isolated and improved transfer deep learning model, Alanazi et al. [48] presented a brain tumor/mass classification framework utilizing MRI. For an unpublished brain MRI dataset, the model attained a high accuracy of 96.89%. Younis et al. [49] evaluated brain tumor diagnosis on a dataset comprising 253 MRI brain images, of which 155 indicated tumors. The algorithm outperformed existing traditional approaches in detecting brain tumors, achieving precision rates of 96, 98.15, and 98.41% and F1 scores of 91.78, 92.6, and 91.29%. In their research, Pedada et al. [50] merged deep learning with computer-aided tumor detection methods, greatly enhancing machine learning. They achieved segmentation accuracies of 93.40 and 92.20%, respectively, using the propsoed U-Net model for brain tumor segmentation (BraTS) Challenge 2017 and 2018 datasets. A deep learning-based classifier named Brain-DeepNet was created by Habibe et al. [51] for the identification and classification of three typical forms of brain malignancies (glioma, meningioma, and pituitary tumors). DNNs trained on labeled OCT images and their ensemble characteristics were proposed in Wang et al. [52] article on “Deep Learning-Based Optical Coherence Tomography Image Analysis for Human Brain Cancer” to combine attenuation and texture data. The model achieved a 96% accuracy in classifying brain tumors. As presented by the studies, deep learning-based CNN architectures are extensively employed in brain tumor diagnosis. he primary goal of this study is to create a useful strategy using MRI scans to detect brain tumors, enabling quick, efficient, and accurate decision-making regarding patients’ conditions.
Zebari et al. [53] devised a deep learning fusion model for brain tumor classification, utilizing data augmentation techniques to overcome the requirement for large training datasets. They extracted deep features from MRI images using VGG16, ResNet50, and convolutional deep belief networks, with Softmax as the classifier. The fusion model, combining features from two DL models, notably enhanced classification accuracy. Azhagiri and Rajesh [54] utilized AlexNet, a deep CNN model, to identify tumors in MRI images. They enhanced AlexNet by adding layers and employed data augmentation techniques to improve accuracy. Their Enhanced AlexNet (EAN) model achieved a remarkable 99.32% accuracy in classifying brain tumors from MRI images, outperforming traditional models. Mandle et al. developed [55] an advanced deep learning model for brain tumor detection, comprising preprocessing, segmentation, and classification stages. They improved image quality using a compound filter and segmented tumors using morphological and threshold-based techniques. The model achieved accurate detection through feature extraction with the grey-level co-occurrence matrix (GLCM) and feature selection optimized by the Whale Social Spider-based Optimization Algorithm (WSSOA), followed by classification using DCNN.
To sum up, these studies collectively underscore the extensive application of CNN and vision transformer models in brain tumor diagnosis, presenting a comprehensive strategy using MRI scans for the detection of brain tumors. This approach enables rapid, efficient, and accurate decision-making regarding patients’ conditions, showcasing the potential of deep learning techniques in significantly advancing the field of medical imaging and diagnosis.
3 Material and methods
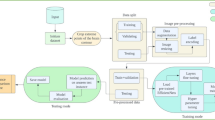
The overall approach, summarized in the diagram of Fig. 2, offers a robust framework consisting of several key components, providing a powerful structure for the identification of brain tumor diseases. We began in Dataset stage by selecting an MRI brain public Dataset [56] was selected from several public datasets and processed in the Pre-processing stage. The images are resized, partitioned, and subjected to basic data augmentation techniques to improve training performance and address class imbalances. The dataset is split into three subsets for deep learning approaches. For small-scale and datasets with limited diversity, data augmentation techniques like flipping, rotation, and shifting are used to increase the diversity of the training data. The next stage, Training models, involves deep learning approaches, specifically utilizing cutting-edge models with ImageNet weights and proposed model. This approach allows for fast convergence and improved performance, especially in small-scale datasets. The models undergo a validation process, where more than 20 deep learning architectures and a total of 45 deep learning models are evaluated for classification. Finally, in the Evaluation stage, models are put to the test on the test data and their performance is evaluated and visually analyzed with Grad-CAM heatmaps [22].
3.1 Dataset
In this study, a large dataset consisting of 7,023 MRI images, including both healthy and diseased samples, was selected from publicly available datasets on Kaggle [56] to effectively train deep learning models. The dataset comprises four distinct classes: Glioma-tumor, Meningioma-tumor, Pituitary-tumor, and No-tumor, with a total of 7,023 brain images with unbalanced classes. The Glioma-tumor class accounted for 1,621 samples, followed closely by Meningioma-tumor with 1,645 samples. The Pituitary-tumor class included 1,757 samples, while the No-tumor class included 2,000 samples. Figure 3 presents randomly selected images from the classes in the utilized dataset.
Gliomas are malignant tumors that occur in a part of the central nervous system. These tumors can originate from brain tissue or cerebrospinal fluid. The majority of brain cancer-related deaths are caused by gliomas, the most aggressive and deadly type of brain tumors [57]. One of the most typical primary brain tumors are gliomas and treating them is still quite difficult [58]. Meningiomas develop from the cells that make up the outer membrane of the brain and spinal cord. Protecting the brain and spinal cord as well as assisting in the movement of the cerebrospinal fluid between the arachnoid and pia layers are the two functions of the meninges, also referred to as the cerebral cortex.
Pituitary tumors, according to a study [2], are tumors that develop as a result of aberrant cell development in the pituitary gland. Although most of these tumors are benign (non-cancerous), they occasionally develop into malignant (cancerous) tumors. Depending on their size, location, and hormone secretion, pituitary tumors can present with a variety of symptoms and indications. The exact causes of pituitary tumors are not yet fully understood, but it is believed that certain factors can increase the risk of tumor formation. No-tumor refers to brain images or situations in which there are no aberrant brain growths or tumors present. In these situations, the brain is thought to be in a healthy and normal state without any signs of cancer or aberrant cell growth.
3.2 Pre-processing
Preprocessing consists of a series of steps to effectively train and evaluate deep learning algorithms. Initially, the raw MRI image is taken and resized from 512 × 512 × 3 pixels to 224 × 224 × 3 pixels to reduce dimension calculations, optimize processing time, and save computational resources. This resizing process is carried out to ensure consistency across the dataset and to facilitate the efficient processing of the deep learning model. The resizing is performed using the bicubic interpolation method. Bicubic interpolation is preferred due to its capability to produce smoother images compared to nearest neighbor and bilinear interpolation methods. This method considers the closest 16 pixels (4 × 4 area) to estimate new pixel values, which helps preserve image quality during the resizing process. This choice is made to maintain the high quality of MRI images because, in medical image processing, this is crucial for accurate tumor detection and classification. In addition, this dimension-reduction process reduces the number of pixels in the image, thereby lightening the computational load and enabling faster results. Next, the image data is shuffled. This step is important to allow the network to work on an irregular dataset during training. The network is prevented from focusing on a particular subset of data by shuffling the images, which also ensures a more representative utilization of the entire dataset [23].
The dataset is divided into four classes that correspond to various forms of brain tumors: glioma, meningioma, pituitary, and no tumor. Each class has a different number of samples, with the “No-tumor” class having the most samples at 2,000. For training, validation, and testing purposes, the dataset is divided into three subsets, as seen in the Table 1.
The meticulous partitioning of the dataset into training, validation, and test sets underpins our method’s reliability and generalizability. This strategic division ensures that our model is trained on a diverse and representative array of MRI scans, encompassing a wide spectrum of tumor types and stages. By validating and testing the model on independent data sets, we are able to rigorously assess its generalization capabilities. This approach is crucial in the medical diagnostics field, where the model’s ability to accurately classify new, unseen patient data is paramount. Such a thorough evaluation framework not only underscores the model’s robustness but also its readiness for deployment in clinical settings, where it can aid in the timely and accurate diagnosis of brain tumors, potentially improving patient outcomes.
Moreover, using the 4,852 samples in the training set, the deep learning model is trained on labeled data to discover patterns and features specific to each tumor class. The model’s hyperparameters are adjusted and its generalizability is checked using the validation set, which has 860 samples. With 1,311 samples, the test set provides an objective assessment of the model’s ultimate performance using entirely new, unseen brain MRI scans. A trustworthy brain tumor classification model may be created with the help of this well-organized dataset, which has a total of 7,023 samples. This could increase diagnostic precision and make it easier for physicians to diagnose brain tumors.
Finally, data augmentation techniques are applied to prevent overfitting and enhance the overall system robustness. Image augmentation increases data diversity by giving the images various transformations, including rotation, flipping, and reflection. This allows the network to work in more varied scenarios and gain a broader perspective on the data. Image augmentation is frequently used to weaken the model and lessen overfitting tendencies. Combining these steps ensures effective image data pre-processing and prepares it for use in the proposed architecture [32].
3.3 Models training
In computer vision, deep learning models have become a ground-breaking innovation [33] that shows considerable potential for classifying and detecting brain tumors. When examining the literature, it becomes evident that CNN models are prominently featured in this context. Based on a thorough review of relevant studies, we carefully selected a set of well-established and widely cutting-edge deep learning architectures for implementation in this research, representing the most popular and up-to-date architectures and effective deep learning structures in brain tumor diagnosis. We selected almost all models belonging to these architectures and also examined the impact of model size on performance. For instance, we not only considered ResNet50 but also included all models ranging from ResNet18 to ResNet101. This approach allowed us to identify successful models for each architecture.”
We chose to use transfer learning since it allows using information learned on previously trained models on big datasets to improve models’ performance on new tasks with small datasets and seeks to speed up model building and boost performance when faced with fresh features. This method is frequently utilized in several disciplines, including sentiment analysis, natural language processing, and computer vision. ImageNet pre-trained models are one of the most used.
DenseNet is a CNN architecture to address the issue of vanishing gradients in deep learning [59]. Each layer within a thick block is connected to all other layers, introducing a novel connection pattern. In addition to making the model more compact and less prone to overfitting, this encourages intensive feature reuse. DenseNet is frequently used in computer vision applications and has demonstrated remarkable performance in a variety of image classification tasks [37].
ResNet, which stands for “Residual Network,” is a CNN design that uses skip connections to overcome the issue of disappearing gradients [60]. These connections enable the network to learn residual functions and facilitate more efficient gradient flow. ResNet has achieved exceptional results in image classification tasks and is widely adopted in computer vision.
EfficientNet is a deep learning architecture introduced by Google that significantly improves image classification performance. It uses a scaling technique called “Compound Scaling” to optimize multiple hyperparameters simultaneously, resulting in higher accuracy with fewer parameters. EfficientNet is particularly suitable for image classification on resource-constrained devices [61].
The Visual Geometry Group at Oxford University created the VGG, a CNN model [40]. Convolutional layers, max pooling, and fully connected layers make up its regular structure. VGG is frequently used as a starting point for creating alternative CNN models because of its remarkable performance on image classification benchmarks like ImageNet [62].
Inception-v4 [63] model expands on the original Inception model using a more CNN design. Inception blocks with parallel convolutional layers, factorized reduction, and stem blocks for initial feature extraction as well as multi-scale feature representation are all included. The model investigates the merging of Inception blocks with residual connections and incorporates auxiliary classifiers for better training.
MobileNetv3 Howard et al. [64] is a CNN architecture designed for low-power devices such as mobile phones. It uses low-dimensional filters during feature extraction to reduce computational costs, making it appropriate for real-time mobile apps. MobileNet performs well in tasks like object detection, face recognition, and image classification.
Xception is a CNN architecture based on the Inception model [65]. It increases accuracy while requiring fewer parameters and calculations by using depthwise separable convolutions and pointwise convolutions. Xception is commonly used in computer vision applications and has demonstrated excellent performance in image classification tasks [45].
MobileViTv2 [66] stands as a cutting-edge lightweight vision transformer model designed specifically for effective image classification and object detection on mobile devices. Its unique attributes include the integration of sophisticated elements like separable self-attention, providing a more computationally efficient alternative to the conventional self-attention mechanism, thereby enhancing accuracy. Additionally, the model improves resource efficiency through layer-wise channel partitioning, which involves the subdivision of each layer into smaller channel groups to reduce memory usage.
InceptionNext [67] represents a CNN architecture that seamlessly integrates the strengths of both the Inception and ConvNeXt architectures. Notable features of InceptionNext include the strategic use of Inception-style modules to decompose computationally expensive large-kernel depthwise convolutions. This decomposition enhances processing efficiency while upholding high accuracy standards. The model also leverages depthwise convolutions adeptly, effectively reducing computational complexity without compromising its feature extraction capabilities.
MaxViT [68] represents an innovative vision transformer model introduced by Google AI researchers in 2022, featuring a distinctive multi-axis attention mechanism that sets it apart in the realm of image classification and object detection. This mechanism operates across multiple spatial dimensions, allowing MaxViT to capture extensive global and contextual information within images, ultimately contributing to superior performance.
Swin Transformer [69], a state-of-the-art vision transformer, excels as a general-purpose backbone for various computer vision tasks, boasting remarkable performance in image classification and dense prediction. Its key features include the construction of hierarchical feature maps for effective capture of local and global information, the use of shifted windows to enhance computational efficiency in self-attention, and a linear computation complexity that enables scalability to high-resolution images.
DeiT III [70], introduced by Facebook Research in 2023, is a state-of-the-art vision transformer designed for efficient image classification on large datasets. Building upon its predecessors, DeiT III features a simplified training recipe based on ResNet-50, reducing complexity, and an efficient data augmentation approach for improved generalization. Employing layer-wise channel partitioning addresses memory consumption, while cross-stage partial connections enhance feature aggregation, contributing to superior performance.
ResMLP [71] is a pioneering image classification architecture based entirely on multi-layer perceptrons (MLPs). ResMLP employs a residual MLP architecture, combining linear layers for independent patch interactions and two-layer feed-forward networks for per-patch channel interactions. The model features an efficient “cross-token attention” mechanism to capture long-range dependencies without the computational complexity of standard self-attention.
GhostNet [72] is a high-performing CNN designed for efficient image classification that the key innovation is the Ghost module, utilizing cost-effective operations like linear transformations and cheap activation functions to enhance feature generation while ensuring computational efficiency. GhostNetV2 [73] represents an enhanced iteration of the GhostNet architecture, aiming to attain superior accuracy and efficiency that presenting advancements in cheap operations complemented by long-range attention mechanisms. RepGhostNet emerges as a novel and hardware-efficient alternative to the Ghost module, a crucial component in the CNN architecture. While maintaining performance to GhostNet, RepGhost achieves a substantial reduction in computational complexity and memory footprint.
FastViT [74], introduced by Apple in 2023, is a groundbreaking CNN-Transformer hybrid architecture designed for efficient image classification and object detection. Utilizing structural reparameterization, it transforms Transformer self-attention into a more computationally efficient convolutional form, significantly reducing complexity while maintaining accuracy. FastViT adopts a hybrid approach, leveraging CNNs for early feature extraction and Transformers for global context aggregation.
CaFormer [75] is a submodel of “MetaFormer Baselines for Vision”, introduced by Weihao Yu et al. in 2023, present a collection of lightweight and efficient transformer architectures tailored for image classification and object detection tasks. Key features include the utilization of efficient token mixers like IdentityFormer, RandFormer, and ConvFormer, reducing computational complexity compared to standard self-attention mechanisms. To address memory consumption, MetaFormer Baselines incorporate layer-wise channel partitioning, dividing layers into smaller channel groups.
3.4 Proposed model
Recent advancements in deep learning have propelled the field of automated brain MRI image diagnosis to new heights. Specifically, incorporating attention mechanisms into CNNs and vision transformer models has markedly improved their performance and efficiency in diagnosing brain conditions. The adoption of the EfficientNetv2 architecture as the foundation for our model is a strategic choice, motivated by its proven efficiency and scalability across a variety of image classification tasks. However, the unique challenges presented by MRI-based brain tumor classification, namely, the high variability in tumor appearances and the subtlety of early manifestations—necessitate an enhancement to this base architecture. To this end, we integrate GAM and ECA into EfficientNetv2, thereby amplifying the model’s capability to discern and prioritize salient features within complex MRI images. This enhancement is not merely an augmentation but a targeted modification designed to elevate the model’s sensitivity to crucial, yet potentially subtle, tumor characteristics, ensuring a significantly improved accuracy in classification tasks. Such an approach is indicative of our method’s robustness, particularly in detecting early-stage tumors where traditional models may falter due to their reliance on more conspicuous tumor features.
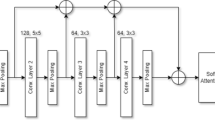
The core of the EfficientNetv2 model includes MBConv and Fused-MBConv layers. Our method enriches this architecture by substituting the SE block with GAM and ECA blocks, in addition to applying strategic rescaling. This refinement aims to forge a model that excels in detecting brain tumors with heightened precision. Figure 4 showcases the model’s architecture, emphasizing the redesigned GAM-enhanced Fused-MBConv and ECA-enhanced MBConv layers tailored for brain tumor identification. This innovative strategy achieves superior classification accuracy with a reduced parameter count when juxtaposed with the conventional EfficientNetv2 model. To offer a thorough understanding of our approach, the study begins with an essential overview of the EfficientNetv2 architecture, then delves into the intricacies of the attention mechanisms, and further investigates the architecture’s scaled modifications.
In this architecture, the MBConv modules, which vary in size and configuration throughout the network, act as advanced convolutional layers designed to boost the network’s ability to learn. They afford the network enhanced flexibility in recognizing more intricate patterns, particularly relevant to visual processing tasks. Meanwhile, the FusedMBConv, an evolved form of the standard MBConv module, aids in the extraction of deeper features through network expansion. This adjustment allows for the development of models that are both efficient and rapid, minimizing the computational demands. In our research, the ECA module is integrated into the MBConv module, substituting the SE module. This enhancement boosts the model’s feature extraction prowess, expanding its learning capabilities while concurrently diminishing the need for computational resources and improving the interaction of depth-wise spatial features. Within the FusedMBConv module, the GAM replaces the SE module. By fostering channel-spatial interactions, GAM retains valuable information, leading to more efficient feature extraction with a reduced number of parameters. Consequently, this approach elevates the model’s overall efficiency while curtailing its computational requirements.
EfficientNetv2 architectures are celebrated for their robust framework, enabling the crafting of scalable and high-efficiency models. The architecture’s scalability is derived from a unique approach that scales the network’s width, depth, and resolution. This method allows EfficientNetv2 to find an equilibrium between the intricacy of the model and its computational demands, ensuring the models are well-suited for both expansive and compact datasets. In this research, we have downscaled the EfficientNetv2’s smallest variant, dubbed EfficientNetv2-Small, to better fit the demands of small-scale brain MRI datasets. This version is characterized by a distinct arrangement of layer repetitions, which indicates the frequency of block repetitions at each network segment. Altering the original layer repetition sequence from 2, 4, 4, 6, 9, 15 to a customized sequence of 2, 3, 3, 4, 6, 12, the model was meticulously adapted for brain MRI data, balancing the model’s size and precision. Such modifications lead to a decrease in the model’s complexity and the total number of parameters, while still preserving its efficacy. This is achieved by employing GAM-enhanced FusedMBConv and ECA-enhanced MBConv blocks, ensuring the model retains high efficiency with minimal impact on accuracy.
3.4.1 EfficientNetv2 architecture
EfficientNetv2, a pioneering CNN-based architecture developed by Tan and Le in 2021, represents a significant advancement in the field, and specifically designed for efficient feature extraction, addressing the shortcomings identified in EfficientNetv1. Initially tailored for optimized binary operations and parameter efficiency, EfficientNetv2 employs Neural Architecture Search (NAS) to uncover an initial architecture that achieves a better balance between accuracy and computational load. Subsequently, the model implements an irregular scaling strategy, progressively adding more layers in later network stages with a designated set of scaling coefficients. Lastly, it enhances model performance and efficiency through progressive learning techniques that systematically increase data regularization and augmentation in line with the growth in image size. Unlike its previous version, EfficientNetv2 extensively utilizes MBConv and Fused-MBConv in the initial layers, opting for a lower MBConv expansion ratio to alleviate the memory access burden. The integration of the FusedMBConv operation, combined with meticulous optimization efforts, positions EfficientNetv2 as a groundbreaking solution in the domain of convolutional classification networks.
3.4.2 Attention mechanism for EfficientNetv2 models
Neural networks can concentrate on the key components of an input by using an attention mechanism. Specifically, it can assist the network in differentiating between background noise and objects of interest in computer vision applications. Utilized extensively to increase CNN models’ accuracy, the attention mechanism has been increasingly popular in recent years. Hard and soft attention are the two different categories of it. Spatial attention, channel attention, and mixed attention are the three main types that have been identified under the soft attention process. To improve its ability to grasp spatial linkages and structures, the network can use spatial attention to selectively focus on spatial regions within the input data. Conversely, channel attention makes it possible for the network to tune feature responses across many channels, making it easier to extract pertinent features while squelching noise. By combining channel and spatial attention, mixed attention gives the network a complete mechanism to simultaneously capture channel-wise and spatial information, enhancing its overall performance in challenging visual recognition tasks. Squeeze and Excitation Block (SE) was one of the first noteworthy attempts to apply channel attention to CNN models. Within the field of CNNs, attention processes have emerged as a keystone for deep learning model development. By emphasizing important properties in a targeted manner, these techniques help the network perform better and have greater discriminative ability. To recalibrate feature responses and enable the network to capture complex interdependencies between various channels, several attention techniques have been developed, such as the Convolutional Block Attention Module (CBAM), the BAM, ECA module, and GAM.
ECA-Net, proposed in 2020 by Wang et al. [43], is an efficient channel attention mechanism for deep CNNs. It captures local cross-channel interactions using a 1D convolution with an adaptive kernel size, replacing traditional fully connected layers. The process involves global average pooling, followed by a 1D convolution and sigmoid activation to modulate the input feature map. Despite notable performance improvements, ECA-Net maintains a lightweight and efficient profile, minimizing model complexity and computational overhead.
CBAM is a lightweight and versatile attention module for CNNs, proposed in 2018 by Sanghyun Woo et al. [76]. It sequentially focuses on both channel and spatial dimensions to enhance feature maps. The channel attention emphasizes important channels via global average pooling and fully connected layers, while spatial attention highlights significant regions through global max pooling and similar layers. The resultant refined feature map is obtained through element-wise multiplication of the two attention maps and is passed to the subsequent CNN layers. The GAM and the CBAM share the same fundamental structure, but they employ different mathematical operators for channel attention and spatial attention modules. Figure 5 illustrates the basic structure of the GAM and CBAM modules.
As shown in Fig. 5, the CBAM significantly boosts the performance of CNNs through the strategic implementation of two consecutive submodules: the Channel Attention Module (CAM) and the Spatial Attention Module (SAM). The CBAM enhances the model’s focus on relevant features by generating two types of attention maps: a one-dimensional channel attention map (Mc) with dimensions of C × 1 × 1, and a two-dimensional spatial attention map (Ms) with dimensions of 1 × H × W. Specifically, Mc is structured to emphasize the importance of each channel across the entire feature map. Conversely, Ms highlights pertinent spatial features within the image. This dual-focus mechanism allows the model to adaptively refine feature representations by prioritizing both channel-wise and spatial information, thereby improving the model’s capability to distinguish significant patterns within the data. Given an intermediate feature map F with dimensions C × H × W as input, CBAM sequentially infers a 1D channel attention map Mc in dimensions of C × 1 × 1, and a 2D spatial attention map Ms in dimensions of 1 × H × W, as demonstrated in Fig. 5. The overall attention process can be formulized in Eq. 1 and Eq. 2.
where, the symbol \(\otimes\) signifies element-wise multiplication. While performing the multiplication, the attention values are copied appropriately: the channel attention values are duplicated across the spatial dimension, and vice versa. \({F}^{{\prime}{\prime}}\) represents the resulting refined output.
In CNNs that utilize attention mechanisms such as ECA, CBAM, and GAM, \({F}_{1}\), \({F}{\prime},\) and \({F}^{{\prime}{\prime}}\) denote different phases in the processing of feature maps. \({F}_{1}\) is derived from the initial convolutional layers and represents the extracted feature set. After undergoing refinement through attention mechanisms, these features are represented as \({F}{\prime}\), where specific elements crucial for tasks like identifying brain tumors are highlighted by adjusting their significance dynamically. \({F}^{{\prime}{\prime}}\) represents a further refinement of \({F}{\prime}\) via additional processing or combining features, thereby boosting the model’s precision and clarity. This enables the model to precisely target and emphasize the most pertinent information in the data. The foundational structure of the CBAM architecture, comprising its two principal components, the CAM and SAM modules, is depicted in Fig. 6.
In the CAM component of the CBAM framework, an attention map focused on channels is generated through leveraging relationships across channel features. This portion of the module prioritizes identifying the key elements in the input. To achieve this, it initially applies average and max pooling to the spatial aspects of the feature maps, producing \({F}_{avg}^{c}\) and \({F}_{max}^{c}\), which correspond to features that have been averaged and maximized across pools, respectively. Following this, these derived values are relayed to a dedicated neural network layer, Mc, which is dimensioned at C × 1 × 1 and contains one hidden layer, where C is the count of channels. The method for calculating the attention dedicated to channels is established through Eq. 3 and Eq. 4.
In this context, σ denotes the sigmoid activation function, while W0 and W1 represent the weight coefficients, and MLP refers to the multi-layer perceptron network.
On the other hand, the SAM crafts a map pinpointing critical spatial regions by analyzing spatial feature correlations. Distinct from its counterpart focusing on channel importance, SAM identifies the specific locations within the input where relevant information is concentrated. For the creation of these spatial attention maps, it conducts average and maximum pooling across the channel dimension, yielding \({{\text{F}}}_{{\text{avg}}}^{{\text{s}}}\in {R}^{1\times H\times W}\) for average-pooled spatial features, and \({{\text{F}}}_{{\text{max}}}^{{\text{s}}}\in {R}^{1\times H\times W}\) for max-pooled spatial features. These outputs are subsequently merged and processed through a convolution layer, represented as \({{\text{M}}}_{\mathrm{s }}\in {{\text{R}}}^{1\times {\text{H}}\times {\text{W}}}\), facilitating the determination of spatial attention through a specialized approach. The computation of spatial attention unfolds in the following Eq. 5 and Eq. 6:
In this description, σ stands for the sigmoid activation function, while f7 × 7 indicates a convolutional operation using a filter with dimensions 7 × 7.
On the other hand, in the domain of brain tumor detection, navigating the dataset’s complexity necessitates a robust mechanism to filter out irrelevant information while emphasizing critical features. The GAM is distinguished by its efficacy in minimizing unnecessary data while accentuating crucial global and dimensional interactions [42]. This superiority of GAM is evident when juxtaposed with other attention mechanisms, primarily due to its advanced capacity for preserving information across the dataset’s intricate variations. GAM advances beyond the foundation laid by the CBAM, preserving the orderly sequence of channel and spatial attentions but with refined submodules for enhanced processing. It employs a 3D permutation technique within the channel attention submodule to maintain comprehensive information across all dimensions. This is followed by the use of a two-layer MLP that intensifies the interplay between channel and spatial elements, ensuring a broad capture of dependencies. The spatial attention component further consolidates this approach by integrating two convolutional layers specifically designed to fuse spatial information more cohesively, as illustrated in Fig. 7.
Operating on the input feature mapping \({F}_{1}\), intermediate state \({F}_{2}\), and final output \({F}_{3}\), as depicted in Eq. 7 and Eq. 8, this mechanism aims to strike a balance between global feature integration and mitigating information loss within the network.
The operational dynamics of GAM are illustrated through equations and visual representations, underlining its structured approach to refining input feature maps. Initially, the channel attention map \({M}_{c}\) is applied to the input feature map \({F}_{1}\), resulting in an intermediate state \({F}_{2}\), which then undergoes further refinement by the spatial attention map \({M}_{s}\), culminating in the final output \({F}_{3}\). This procedural flow ensures a comprehensive enhancement of significant features within the images, making GAM an invaluable tool in the nuanced field of medical image analysis and brain tumor detection.
3.5 Evaluation metrics
Different performance metrics are used in machine learning to assess the efficacy of created models or algorithms, notably in classifying and detecting brain tumors. Accuracy, Precision, Recall, F1-score, Sensitivity, and Specificity are some of these measurements. The F1-score provides a balanced measurement that takes Precision and recall into account. Sensitivity emphasizes reducing false negatives, while specificity assesses the accuracy of negative predictions. Accuracy measures the overall correctness of predictions, while Precision focuses on the accuracy of positive predictions, Recall evaluates the model’s capacity to identify positive cases correctly. Researchers can learn more about the model’s effectiveness and capacity to correctly categorize cases of brain tumors by using these measures and examining the confusion matrix produced. These metrics can be computed using the following formulas:
4 Discussion and results
In this section, the experimental results are meticulously analyzed. A description of the experimental setup, elucidating how the experimental process was conducted, precedes a comprehensive discussion alongside tables and graphs presenting the outcomes of deep learning architectures. Furthermore, the interpretability of CNN architectures is demonstrated through the extraction of heat map regions of brain tumor areas using Grad-CAM. Finally, a detailed comparison with state-of-the-art (SOTA) models is provided to evaluate the proposed method against existing benchmarks.
4.1 Experimental design
The computer utilized in the experiments had the following hardware configuration: a Linux-based operating system called Ubuntu 21.04 was employed; an NVIDIA RTX 2080TI graphics card with 11 GB of GDDR6 RAM and 4352 CUDA cores was part of the GPU hardware; an Intel Core i9 9900X with 10 cores, 3.50 GHz, and a 19.25 MB Intel® Smart Cache served as the processor and 32 GB of DDR4 RAM. Python was utilized as the programming language, and the PyTorch deep learning library was used.
4.2 Results
This section provides a comprehensive assessment of advanced deep learning models in the context of brain tumor diagnosis. It discusses the performance metrics and the outcomes depicted in the confusion matrix associated with the proposed diagnostic model. The findings presented in Table 2 demonstrate that, with the exception of VGG19, all deep learning models achieve accuracy rates exceeding 99% in correctly identifying brain tumors. These results underline the efficacy and reliability of deep learning methodologies in aiding medical professionals in accurately diagnosing brain tumors. They highlight the potential of these models to significantly improve diagnostic accuracy and patient outcomes in the field of neurology.
Table 2 compares the performance of various deep learning architectures in classifying MRI images for brain tumor identification. It evaluates popular models like VGG, ResNet, DenseNet, Inception, MobileNet, MobileViTv2, FastViT, GhostNet, RepGhostNet, MaxViT, Swin, DeiT3, ResMLP, and Caformer based on crucial performance metrics such as accuracy, precision, sensitivity, and F1 score.
A key finding is the Proposed Model’s demonstration of the highest performance with an accuracy of 99.77, precision of 99.76, sensitivity of 99.75, and an F1 score of 99.75%. This indicates a significant superiority of the Proposed Model over all other deep learning models examined for brain tumor classification, highlighting its ability to perform more precise and effective classification thanks to specialized attention mechanisms and model optimizations used particularly for detecting brain tumors.
Furthermore, architectures like DenseNet-121 and Inceptionv3 also showcase remarkable performance with accuracy rates around 99.47%, while the VGG-19 model shows a relatively lower performance at 98.55%. This variance in performance indicates how the architecture of the model and the feature extraction techniques used can significantly affect outcomes. Notably, models such as FastViT-s12 and RepGhostNet-200, displaying high accuracy rates of 99.54%, underscore the potential of transformer-based and lightweight models in visual classification tasks. These results demonstrate how various deep learning architectures can perform differently in specific medical imaging tasks like brain tumor diagnosis and the importance of selecting the most appropriate model.
Similarly, notable architectures like MaxViT-base, Swin-base, DeiT3-base, and ResMLP-24 have shown impressive performance in brain tumor identification tasks. Particularly, MaxViT-base and Swin-base stand out with accuracy rates of 99.54 and 99.47%, respectively, showcasing their capability to effectively process complex visual features and their significant potential in deep learning applications for medical imaging. DeiT3-base emphasizes the strength of transformer architectures with an accuracy of 99.31%, while ResMLP-24 validates the efficacy of MLP architectures in this field with an accuracy of 99.39%. In conclusion, these findings illuminate the efficacy and potential of deep learning approaches in medical imaging, especially in diagnosing brain tumors using MRI. Figure 8 shows a bar graph for all models mentioned in Table 2.
In Fig. 8, the premier performing model, referred to as the Proposed Model, is distinguished in red, while the subsequent top four models, namely Caformer-s36, MaxViT-base, RepGhostNet-200, and FastViT-s12, are presented in green. The remaining models are shaded in gray to indicate their comparatively lower performance. Furthermore, Table 3 presents a comparison of the model performances within the EfficientNetv1 and EfficientNetv2 frameworks using brain MRI datasets. It also highlights the influence of employing different attention mechanisms for CNN-based architectures in conjunction with the implementation of the EfficientNetv2-small model, alongside the outcomes achieved by the Proposed Model.
Table 3 provides a comprehensive comparison between various EfficientNet models used for detecting brain tumors and a proposed model. This comparison offers a detailed analysis in terms of performance metrics such as accuracy, precision, recall, and F1-score, while also aiming to deeply investigate the impact of attention mechanisms. This section presents analyses on the performance of both the EfficientNet and EfficientNetv2 series, along with the proposed model (GAM + ECA), and the effects of attention mechanisms on brain tumor detection. A thorough comparison between the EfficientNet and EfficientNetv2 series demonstrates that both series deliver excellent performance metrics across various configurations, which is illustrated in Fig. 9.
Figure 9 illustrates the achievements on the accuracy metric achieved by EfficientNet models and EfficientNet models with added attention mechanisms. The proposed model demonstrates the highest achievement (highlighted in red), while the closest 4 models are also highlighted in green, and models and optimizations belonging to other EfficientNet architectures are shown in gray.
Notably, the EfficientNetv2 series and its attention mechanism-enhanced variants stand out for improving performance significantly without increasing model complexity. The efficacy of the proposed model is concretized through the integration of different attention mechanisms into the EfficientNetv2-Small model. This integration, which replaces the SE modules in FusedMBConv blocks with GAM and substitutes SE modules in MBConv blocks with ECA mechanisms, represents a hybrid approach. This strategy reduces the total parameter count of the model to 14.41 million while elevating the accuracy to 99.77, precision to 99.76, recall to 99.75, and F1-score to 99.75%. This highlights the model’s lightweight structure and its capability to achieve superior performance metrics while reducing computation resources and training time requirements. Notably, this optimization has also resulted in significant reductions in GPU usage and FLOPs.
The proposed model’s efficiency is evident in its reduced parameter count and computational footprint. Specifically, the model leverages depth-wise separable convolutions and a scalable architecture that adjusts the model’s width, depth, and resolution in a compound manner, optimizing for speed and memory usage without compromising performance. Compared to traditional CNN architectures such as VGG and ResNet, the proposed model demonstrates significant improvements in computational speed and memory efficiency. For instance, while VGG-16 may require over 138 million parameters, the proposed EfficientNetv2-based model, even with the added attention mechanisms, maintains a parameter count significantly lower, contributing to faster training times and reduced memory requirements. In terms of inference speed, the proposed model exhibits a marked improvement over classical models like VGG-16 and ResNet-50. Benchmarked on the same hardware, the proposed model achieves inference times up to 2 × faster than VGG-16 and 1.5 × faster than ResNet-50, facilitating real-time analysis and classification of MRI images.
The analysis concerning parameter count and computational cost (FLOPs, GPU usage) reveals that a higher parameter counts or computational intensity does not necessarily correlate with better performance. EfficientNet-B0 and EfficientNet-B1 models, with 5.3 and 7.8 million parameters respectively, offer accuracy rates of 99.31 and 99.47%. In contrast, EfficientNet-B3 and EfficientNet-B4 models, with respectively 12 and 19 million parameters, maintain a similar high level of accuracy at 99.54% along with comparable high precision, recall, and F1-scores. The largest model in the EfficientNetv2 series, EfficientNetv2-Large, despite having 48 million parameters, showcases impressive performance metrics with an accuracy of 99.54, precision of 99.53, recall of 99.50, and F1-score of 99.52%.
Investigations into the effect of attention mechanisms show that the integration of ECA into the EfficientNetv2-Small model reduces the parameter count from 20.19 to 17.76 million, yielding significant improvements in accuracy (to 99.62), precision (to 99.61), recall (to 99.59), and F1-score (to 99.60%). The use of GAM, while increasing the parameter count to 88.67 million, elevates the accuracy from 99.54 to 99.69%, significantly enhancing overall performance and yielding positive outcomes in other metrics. However, the integration of CBAM increases the parameter count to 22.52 million without significant improvements in other metrics. The proposed model (GAM + ECA) demonstrates not only a reduction in parameter count and computational cost but also significant improvements in critical performance metrics such as accuracy, precision, recall, and F1-score when compared to existing models. This evidences significant progress in deep learning-based detection of brain tumors, proving the model’s ability to strike an effective balance between performance and efficiency. The success of the proposed model underscores the importance of strategic integration of attention mechanisms in developing deep learning models. These findings lay a foundation for future research, encouraging the development of more efficient and effective deep learning models. Figure 10 displays the confusion matrix depicting the class performance of the proposed model.
Upon reviewing Fig. 10, the detailed evaluation of the confusion matrix confirms the classification model’s superior performance in differentiating among four categories: Glioma, Meningioma, Pituitary, and No-tumor. The model exhibits impeccable precision in identifying Glioma, with 299 correct predictions, and Pituitary tumors, with 298 correct predictions, underscoring near-flawless accuracy. It similarly achieves outstanding accuracy in recognizing Meningioma and No-tumor cases, with 306 and 405 correct predictions respectively. Discrepancies were minimal, involving one Meningioma case misclassified as Glioma, and two instances where the model incorrectly labeled Pituitary tumors as Meningioma. These results suggest that the model secures high metrics in precision, recall, and F1-scores for each category, reflecting a well-balanced and sturdy capacity to distinguish specific conditions as well as general non-tumor instances. The scant incidence of misclassifications emphasizes the model’s adeptness at discerning between various brain tumor types and its strong aptitude for correctly identifying non-tumor cases, which serves to underline both its high rate of accuracy and its robust generalization capability across distinct tumor classes.
4.3 Visualization analysis using grad-CAM
Grad-CAM, which stands for Gradient-weighted Class Activation Mapping, is an influential technique employed to interpret and visualize how CNNs make decisions. Its main objective is to pinpoint the significant regions in an input image that greatly influence the network’s predictions for a specific class. In the realm of brain tumor classification, Grad-CAM serves as a pivotal tool, visually illustrating the specific features that CNN-based models prioritize. This elucidation offers invaluable insights into the rationale behind the model’s classification decisions, thereby bolstering our understanding of its diagnostic capabilities. Through this analysis, crucial for precise brain tumor identification and classification within medical imaging contexts, the model’s credibility is significantly enhanced. Empirical evidence corroborates that the Proposed Model excels in accurately classifying brain tumors, underscoring its superiority in comparison to other models. The integration of Grad-CAM into our model addresses a critical need for interpretability in AI-driven medical diagnostics. This technique illuminates the model’s decision-making process by highlighting the specific regions within MRI scans that influence its classification decisions. In the context of brain tumor diagnosis, where understanding the basis of a model’s prediction is as vital as the prediction’s accuracy, Grad-CAM serves as an invaluable tool. It not only aids radiologists and clinicians in validating the AI’s findings but also fosters a deeper trust in the technology. By providing transparent and interpretable insights into the model’s operational logic, Grad-CAM enhances the collaborative potential between AI models and medical professionals, paving the way for more informed and effective treatment planning. Figure 11 illustrates how the integration of Grad-CAM with the Proposed Model focuses on particular locations in brain tumor regions.
As seen in Fig. 11, the two rows that are visible offer important details on the interpretability of artificial intelligence and the accuracy of the model for detecting brain tumors. The radiologist-marked brain tumor locations in the first row serve as manually positioned reference points. The Proposed Model’s integration with Grad-CAM yields heats map visualizations in the second row that reveal the underlying causes of the model’s categorization choices. The integration of the Proposed Model with Grad-CAM clearly shows the tumor locations that the model concentrates on during brain tumor diagnosis. Nearly all of the images feature heat map representations illustrating the locations where the model focusses its attention, successfully identifying tumor regions. This study not only strengthens the model’s trustworthiness but also acts as an essential tool for pinpointing the causes of incorrect classifications.
Additionally, the conclusions made by the model are interpretable, which promotes confidence among radiologists and specialists and allows for improved therapeutic decision-making. Grad-CAM, in this context, emerges as an important tool that visually interprets the operation of the Proposed Model and identifies tumor regions, supporting the interpretability of artificial intelligence in crucial areas like the classification of brain tumors. Grad-CAM visualizations for Proposed Model with some mis-regions and misclassifications are shown in Fig. 12.
In Fig. 12, the Grad-CAM analysis results of Proposed Model include some misclassifications and inaccurate focus areas. Upon examination, we can observe that the integration of Proposed Model with Grad-CAM fails to fully characterize misclassified brain tumors and focuses on incorrect regions. Both the erroneous detections of Proposed Model and Grad-CAM’s focus on areas other than misclassified tumor regions demonstrate the compatibility and effectiveness of both methods, showcasing Grad-CAM’s successful results in CNN-based deep learning techniques.
Regarding tumor-free images, models using the Grad-CAM technique shift their focus to different areas. Despite achieving high accuracy, it can be said that Grad-CAM does not always provide precise localization. This highlights the need for developing more robust and advanced techniques in artificial intelligence and medical image processing to enhance interpretability beyond Grad-CAM’s limitations.
As observed in Fig. 7 and Fig. 8, Grad-CAM can help overcome some of the “black box” nature of CNN models and make their decision mechanisms more interpretable. As demonstrated by the Proposed Model, this method can enhance confidence in the model’s medical diagnoses and enable doctors to be more involved in the validation and interpretation of the model’s diagnoses. However, it cannot be claimed to provide complete interpretability. Nevertheless, by developing advanced techniques that address the challenges in this regard, artificial intelligence can achieve a higher level of interpretability in the medical field.
4.4 Comparison with cutting-edge methods
To validate the superiority of our proposed model, we conducted extensive experiments and benchmarked its performance against a comprehensive suite of state-of-the-art deep learning models. This comparative analysis is pivotal, as it situates our model within the current landscape of MRI-based brain tumor classification technologies, demonstrating its efficacy and efficiency. Table 4 displays our proposed model’s superior performance to other cutting-edge methods.
Considering Table 4, our Proposed Model has achieved a significant success in MRI-based brain tumor classification, reaching an impressive accuracy rate of 99.76%. This accomplishment demonstrates a notable superiority over traditional CNN architectures or hybrid approaches (CNN + SVM, DNN + SVM) utilized by Celik and Inik [31], Anantharajan et al. [46], and Deepak and Ameer [81], which offer accuracy rates ranging from 95.60 to 97.93%. Our model, showcasing how attention mechanisms can enhance classification precision, surpasses these rates by a considerable margin.
Additionally, studies employing ensemble methods and advanced CNN frameworks, such as those by Remzan et al. [47] and Rahman and Islam [78], have shown accuracy rates below 98.20%. These innovative approaches have been outperformed by the efficiency and accuracy of our EfficientNetv2-based model integrated with attention mechanisms. In addition, while Tabatabaei et al. [80] proposed a model incorporating attention mechanisms and achieved 99.30% accuracy on the Figshare dataset, our model has further advanced this accuracy, particularly emphasizing the effectiveness of specific attention mechanisms such as GAM and ECA.
The superiority of our model extends beyond a single dataset, as it demonstrates higher accuracy rates compared to models reported by Zebari et al. [53], Muezzinoglu et al. [79], and others on different datasets, proving the robustness and generalizability of our approach. The standout aspect of our model lies in the integration of attention mechanisms such as GAM and ECA with EfficientNetv2. This specific combination enables the model to accurately focus on significant features in complex MRI images, thus facilitating its superior performance.
Furthermore, The Kaggle dataset, one of the largest datasets in brain tumor identification comparisons, introduces a higher level of complexity to the task. The use of the proposed model’s training-validation-test split (unlike many studies which only utilize train-validation or cross-validation) is particularly notable. This approach aligns well with best practices for deep learning models in the challenging field of brain tumor identification, allowing for the generalization of these models over unseen test data. The exceptional performance of our model, underscored by a high-test accuracy and supported by a robust validation framework, sets a new benchmark in the field. By rigorously comparing our model against existing solutions, we provide a clear and compelling case for its adoption in clinical settings, where it can significantly enhance the accuracy and efficiency of brain tumor diagnoses. This comparative analysis not only establishes our model’s technical excellence but also its potential to contribute meaningfully to the advancement of medical imaging analysis, offering a promising avenue for future research and application.
4.5 Limitations and future directions
The proposed approach, enhancing EfficientNetv2 with attention mechanisms, presents a promising advancement in MRI-based brain tumor classification, spanning from clinical diagnostics to research and development. Specifically, this model holds the potential to significantly enhance the accuracy and efficiency of brain tumor identification in clinical diagnostics, aiding radiologists in making faster and more reliable decisions. Its application extends to providing personalized and effective treatment strategies, leveraging detailed information about tumor characteristics provided by the model. Beyond healthcare, this approach could serve as a valuable tool in medical education by offering a practical example of applying advanced deep learning techniques in medical imaging analysis.
However, despite its promising applications, the model faces limitations that need to be addressed to fully realize its potential. One significant constraint is the size and diversity of the dataset, which impacts the model’s ability to generalize across a wide range of brain tumors accurately. Additionally, while the integration of Grad-CAM enhances model interpretability, its opacity at the decision-making process level may hinder trust and acceptance among healthcare professionals. Computational requirements for processing comprehensive datasets or integrating the model into real-time diagnostic platforms could pose challenges, especially in resource-constrained environments.
Looking ahead, various avenues in research and development offer opportunities to overcome these limitations and expand the model’s applicability. Efforts to enhance dataset representation through large-scale data from different sources can reduce biases and enhance the model’s robustness. Advanced interpretability techniques beyond Grad-CAM could make the model’s inner workings more transparent and understandable for end-users. Optimizing the model for computational efficiency could facilitate its deployment across various platforms, including mobile and edge computing devices, making advanced diagnostics more accessible. In conclusion, while the proposed approach represents a significant step forward in applying deep learning to medical imaging, addressing current limitations and exploring future directions are crucial for its evolution. Progress in dataset diversity, interpretability, computational efficiency, and integration with broader healthcare data will unlock its full potential, paving the way for more complex, accessible, and personalized medical diagnostics and treatment planning.
5 Conclusion
This study presents an innovative adaptation of the EfficientNetv2 architecture enhanced and re-scaled with Global Attention Mechanism (GAM) and Efficient Channel Attention (ECA) for accurate classification of MRI-based brain tumors. Through extensive experiments, our model has set a new benchmark in the field, achieving an exceptional test accuracy of 99.76%. The integration of attention mechanisms significantly enhances the model’s focus on important features in MRI images, leading to superior performance in detecting a wide range of brain tumors. Moreover, the study explores the performance of various attention mechanisms and 45 deep learning models in brain tumor classification. Additionally, the application of Grad-CAM visualization techniques provides valuable interpretability into the model’s decision-making process, offering crucial insights for clinical evaluation. This advancement not only propels the field of medical imaging analysis forward but also underscores the critical role of attention mechanisms in improving the accuracy and interpretability of deep learning models in brain tumor diagnosis, promising to enhance patient care and treatment outcomes.
References
Siegel, R.L., Giaquinto, A.N., Jemal, A.: Cancer statistics, 2024. CA Cancer J. Clin. 74(1), 12–49 (2024). https://doi.org/10.3322/caac.21820
Herholz, K., Langen, K.J., Schiepers, C., Mountz, J.M.: Brain tumors. Semin. Nucl. Med. 42, 356–370 (2012). https://doi.org/10.1053/j.semnuclmed.2012.06.001
Bondy, M.L., Scheurer, M.E., Malmer, B., Barnholtz-Sloan, J.S., Davis, F.G., Il’yasova, D., Kruchko, C., McCarthy, B.J., Rajaraman, P., Schwartzbaum, J.A., Sadetzki, S., Schlehofer, B., Tihan, T., Wiemels, J.L., Wrensch, M., Buffler, P.A.: Brain tumor epidemiology: consensus from the brain tumor epidemiology consortium. Cancer 113(7 Suppl), 1953–1968 (2008)
Lamba, N., Wen, P.Y., Aizer, A.A.: Epidemiology of brain metastases and leptomeningeal disease. Neuro Oncol. 23, 1447–1456 (2021). https://doi.org/10.1093/neuonc/noab101
Roth, H.R., Lu, L., Liu, J., Yao, J., Seff, A., Cherry, K., Kim, L., Summers, R.M.: Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging 35, 1170–1181 (2016). https://doi.org/10.1109/TMI.2015.2482920
Siegel, R.L., Miller, K.D., Wagle, N.S., Jemal, A.: Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023). https://doi.org/10.3322/CAAC.21763
Charles, N.A., Holland, E.C., Gilbertson, R., Glass, R., Kettenmann, H.: The brain tumor microenvironment. Glia 59, 1169–1180 (2011). https://doi.org/10.1002/glia.21136
Villanueva-Meyer, J.E., Mabray, M.C., Cha, S.: Current clinical brain tumor imaging. Clin. Neurosurg. 81, 397–415 (2017). https://doi.org/10.1093/neuros/nyx103
Liu, Z., Tong, L., Chen, L., Jiang, Z., Zhou, F., Zhang, Q., Zhang, X., Jin, Y., Zhou, H.: Deep learning based brain tumor segmentation: a survey. Complex and Intelligent Systems. 9, 1001–1026 (2023). https://doi.org/10.1007/s40747-022-00815-5
Pacal, I.: A novel swin transformer approach utilizing residual multi-layer perceptron for diagnosing brain tumors in MRI images. Int. J. Mach. Learn. Cybern. (2024). https://doi.org/10.1007/s13042-024-02110-w
Leblebicioglu, I., Lubbad, M., Yilmaz, O.M.D., Kilic, K., Karaboga, D., Basturk, A., Akay, B., Nalbantoglu, U., Yilmaz, S., Ayata, M., Pacal, I.: A robust deep learning model for the classification of dental implant brands. J. Stomatol. Oral Maxillofac. Surg. 8, 101818 (2024). https://doi.org/10.1016/j.jormas.2024.101818
Farooq, M.U., Ullah, Z., Khan, A., Gwak, J.: DC-AAE: dual channel adversarial autoencoder with multitask learning for KL-grade classification in knee radiographs. Comput. Biol. Med. 167, 107570 (2023). https://doi.org/10.1016/j.compbiomed.2023.107570
Zador, A.M.: A critique of pure learning and what artificial neural networks can learn from animal brains. Nat. Commun. 10(1), 3770 (2019). https://doi.org/10.1038/s41467-019-11786-6
Myers, D., Mohawesh, R., Chellaboina, V.I., Sathvik, A.L., Venkatesh, P., Ho, Y.H., Henshaw, H., Alhawawreh, M., Berdik, D., Jararweh, Y.: Foundation and large language models: fundamentals, challenges, opportunities, and social impacts. Cluster. Comput. 27, 1–26 (2024)
Lubbad, M., Karaboga, D., Basturk, A., Akay, B., Nalbantoglu, U., Pacal, I.: Machine learning applications in detection and diagnosis of urology cancers: a systematic literature review. Neural Comput. Appl. 36, 6355–6379 (2024). https://doi.org/10.1007/s00521-023-09375-2
Ullah, Z., Usman, M., Latif, S., Gwak, J.: Densely attention mechanism based network for COVID-19 detection in chest X-rays. Sci. Rep. 13, 261 (2023). https://doi.org/10.1038/s41598-022-27266-9
Kunduracioglu, I., Pacal, I.: Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J. Plant Dis. Prot. (2024). https://doi.org/10.1007/s41348-024-00896-z
Ullah, Z., Usman, M., Gwak, J.: MTSS-AAE: multi-task semi-supervised adversarial autoencoding for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 216, 119475 (2023). https://doi.org/10.1016/j.eswa.2022.119475
Karaman, A., Pacal, I., Basturk, A., Akay, B., Nalbantoglu, U., Coskun, S., Sahin, O., Karaboga, D.: Robust real-time polyp detection system design based on YOLO algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (ABC). Expert Syst. Appl. 221, 119741 (2023). https://doi.org/10.1016/j.eswa.2023.119741
Kolides, A., Nawaz, A., Rathor, A., Beeman, D., Hashmi, M., Fatima, S., Berdik, D., Al-Ayyoub, M., Jararweh, Y.: Artificial intelligence foundation and pre-trained models: fundamentals, applications, opportunities, and social impacts. Simul. Model. Pract. Theory 126, 102754 (2023). https://doi.org/10.1016/j.simpat.2023.102754
Ullah, Z., Usman, M., Latif, S., Khan, A., Gwak, J.: SSMD-UNet: semi-supervised multi-task decoders network for diabetic retinopathy segmentation. Sci. Rep. 13, 9087 (2023). https://doi.org/10.1038/s41598-023-36311-0
Lubbad, M.A.H., Kurtulus, I.L., Karaboga, D., Kilic, K., Basturk, A., Akay, B., Nalbantoglu, O.U., Yilmaz, O.M.D., Ayata, M., Yilmaz, S., Pacal, I.: A comparative analysis of deep learning-based approaches for classifying dental implants decision support system. Journal of Imaging Informatics in Medicine. (2024). https://doi.org/10.1007/s10278-024-01086-x
Lecun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015). https://doi.org/10.1038/nature14539
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017). https://doi.org/10.1145/3065386
Pacal, I.: Enhancing crop productivity and sustainability through disease identification in maize leaves: exploiting a large dataset with an advanced vision transformer model. Expert Syst. Appl. 238, 122099 (2024). https://doi.org/10.1016/j.eswa.2023.122099
Celard, P., Iglesias, E.L., Sorribes-Fdez, J.M., Romero, R., Vieira, A.S., Borrajo, L.: A survey on deep learning applied to medical images: from simple artificial neural networks to generative models 35, 2291–2323 (2023)
Wang, J., Zhu, H., Wang, S.H., Zhang, Y.D.: A review of deep learning on medical image analysis. Mobile Networks and Applications. 26, 351–380 (2021). https://doi.org/10.1007/s11036-020-01672-7
Pacal, I., Kılıcarslan, S.: Deep learning-based approaches for robust classification of cervical cancer. Neural Comput. Appl. 35, 18813–18828 (2023). https://doi.org/10.1007/s00521-023-08757-w
Karaman, A., Karaboga, D., Pacal, I., Akay, B., Basturk, A., Nalbantoglu, U., Coskun, S., Sahin, O.: Hyper-parameter optimization of deep learning architectures using artificial bee colony (ABC) algorithm for high performance real-time automatic colorectal cancer (CRC) polyp detection. Appl. Intell. (2022). https://doi.org/10.1007/s10489-022-04299-1
Pacal, I., Karaboga, D.: A robust real-time deep learning based automatic polyp detection system. Comput. Biol. Med. 134, 104519 (2021). https://doi.org/10.1016/J.COMPBIOMED.2021.104519
Celik, M., Inik, O.: Development of hybrid models based on deep learning and optimized machine learning algorithms for brain tumor multi-classification. Expert Syst. Appl. 238, 122159 (2024). https://doi.org/10.1016/j.eswa.2023.122159
Pacal, I.: MaxCerVixT: a novel lightweight vision transformer-based approach for precise cervical cancer detection. Knowl Based Syst. 289, 111482 (2024). https://doi.org/10.1016/j.knosys.2024.111482
Nazir, M., Shakil, S., Khurshid, K.: Role of deep learning in brain tumor detection and classification (2015 to 2020): a review. Comput. Med. Imaging Graph. 91, 101940 (2021). https://doi.org/10.1016/j.compmedimag.2021.101940
Jyothi, P., Singh, A.R.: Deep learning models and traditional automated techniques for brain tumor segmentation in MRI: a review. Artif. Intell. Rev. 56, 2923–2969 (2023). https://doi.org/10.1007/s10462-022-10245-x
Chahal, P.K., Pandey, S., Goel, S.: A survey on brain tumor detection techniques for MR images. Multimed Tools Appl. 79, 21771–21814 (2020). https://doi.org/10.1007/s11042-020-08898-3
Bhagyalaxmi, K., Dwarakanath, B., Reddy, P.V.P.: Deep learning for multi-grade brain tumor detection and classification: a prospective survey. Multimed Tools Appl. (2024). https://doi.org/10.1007/s11042-024-18129-8
Ali, S., Li, J., Pei, Y., Khurram, R., Rehman, K., Mahmood, T.: A comprehensive survey on brain tumor diagnosis using deep learning and emerging hybrid techniques with multi-modal MR image 29, 4871–4896 (2022)
Farajzadeh, N., Sadeghzadeh, N., Hashemzadeh, M.: Brain tumor segmentation and classification on MRI via deep hybrid representation learning. Expert Syst. Appl. 224, 119963 (2023). https://doi.org/10.1016/j.eswa.2023.119963
Awuah, W.A., Adebusoye, F.T., Wellington, J., David, L., Salam, A., Weng Yee, A.L., Lansiaux, E., Yarlagadda, R., Garg, T., Abdul-Rahman, T., Kalmanovich, J., Miteu, G.D., Kundu, M., Mykolaivna, N.I.: Recent outcomes and challenges of artificial intelligence, machine learning and deep learning applications in neurosurgery – Review applications of artificial intelligence in neurosurgery. World Neurosurg X. 23, 100301 (2024). https://doi.org/10.1016/j.wnsx.2024.100301
Levy, A.S., Bhatia, S., Merenzon, M.A., Andryski, A.L., Rivera, C.A., Daggubati, L.C., Di, L., Shah, A.H., Komotar, R.J., Ivan, M.E.: Exploring the Landscape of Machine Learning Applications in Neurosurgery: A Bibliometric Analysis and Narrative Review of Trends and Future Directions 181, 108–115 (2024)
Tan, M., Le, Q.V.: EfficientNetV2: smaller models and faster training 139, 10096–10106 (2021)
Y Liu Z Shao N 2021 Hoffmann Global Attention Mechanism: Retain Information to Enhance Channel-Spatial Interactions.
Q Wang B Wu P Zhu P Li W Zuo Q Hu 2019 ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks
RR Selvaraju M Cogswell A Das R Vedantam D Parikh D 2017 Batra Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 618–626. Institute of Electrical and Electronics Engineers Inc.
Ullah, Z., Usman, M., Jeon, M., Gwak, J.: Cascade multiscale residual attention CNNs with adaptive ROI for automatic brain tumor segmentation. Inf Sci (N Y). 608, 1541–1556 (2022). https://doi.org/10.1016/j.ins.2022.07.044
Anantharajan, S., Gunasekaran, S., Subramanian, T.: MRI brain rain tumor detection using deep learning and machine learning approaches. Measurement: Sensors. 31, 101026 (2024). https://doi.org/10.1016/j.measen.2024.101026
Remzan, N., Tahiry, K., Farchi, A.: Advancing brain tumor classification accuracy through deep learning: harnessing radimagenet pre-trained convolutional neural networks, ensemble learning, and machine learning classifiers on MRI brain images. Multimed Tools Appl. (2024). https://doi.org/10.1007/s11042-024-18780-1
Alanazi, M.F., Ali, M.U., Hussain, S.J., Zafar, A., Mohatram, M., Irfan, M., Alruwaili, R., Alruwaili, M., Ali, N.H., Albarrak, A.M.: Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors. 22, 372 (2022). https://doi.org/10.3390/s22010372
Younis, A., Qiang, L., Nyatega, C.O., Adamu, M.J., Kawuwa, H.B.: Brain tumor analysis using deep learning and VGG-16 ensembling learning approaches. Applied Sciences (Switzerland) 12, 7282 (2022). https://doi.org/10.3390/app12147282
Pedada, K.R., Bhujanga Rao, A., Patro, K.K., Allam, J.P., Jamjoom, M.M., Samee, N.A.: A novel approach for brain tumour detection using deep learning based technique. Biomed. Signal Process. Control 82, 104549 (2023). https://doi.org/10.1016/j.bspc.2022.104549
Habiba, S.U., Islam, M.K., Nahar, L., Tasnim, F., Hossain, M.S., Andersson, K.: Lecture Notes in Networks and Systems. 569, 550–560 (2023). https://doi.org/10.1007/978-3-031-19958-5_52/COVER
Wang, N., Lee, C.-Y., Park, H.-C., Nauen, D.W., Chaichana, K.L., Quinones-Hinojosa, A., Bettegowda, C., Li, X.: Deep learning-based optical coherence tomography image analysis of human brain cancer. Biomed. Opt. Express 14, 81 (2023). https://doi.org/10.1364/boe.477311
Zebari, N.A., Mohammed, C.N., Zebari, D.A., Mohammed, M.A., Zeebaree, D.Q., Marhoon, H.A., Abdulkareem, K.H., Kadry, S., Viriyasitavat, W., Nedoma, J., Martinek, R.: A deep learning fusion model for accurate classification of brain tumours in magnetic resonance images. CAAI Trans Intell Technol. (2024). https://doi.org/10.1049/cit2.12276
Azhagiri, M., Rajesh, P.: EAN: enhanced AlexNet deep learning model to detect brain tumor using magnetic resonance images. Multimed Tools Appl. (2024). https://doi.org/10.1007/s11042-024-18143-w
Mandle, A.K., Sahu, S.P., Gupta, G.P.: WSSOA: whale social spider optimization algorithm for brain tumor classification using deep learning technique. International Journal of Information Technology (Singapore). (2024). https://doi.org/10.1007/s41870-024-01782-5
Brain Tumor MRI Dataset | Kaggle, https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset?select=Training
Ostrom, Q.T., Barnholtz-Sloan, J.S.: Current state of our knowledge on brain tumor epidemiology. Curr. Neurol. Neurosci. Rep. 11, 329–335 (2011). https://doi.org/10.1007/s11910-011-0189-8
Tummala, S., Kadry, S., Bukhari, S.A.C., Rauf, H.T.: Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Curr. Oncol. 29, 7498–7511 (2022). https://doi.org/10.3390/curroncol29100590
G Huang Z Liu L van der Maaten KQ Weinberger 2016 Densely Connected convolutional networks
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016, 770–778 (2016). https://doi.org/10.1109/CVPR.2016.90
M Tan QV Le 2019 EfficientNet: rethinking model scaling for convolutional neural networks
K Simonyan 2015 A Zisserman Very deep convolutional networks for large-scale image recognition. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. 1–14
C Szegedy S Ioffe V Vanhoucke AA Alemi 2016 Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. 31st AAAI Conference on Artificial Intelligence, AAAI 2017. 4278–4284. https://doi.org/10.1609/aaai.v31i1.11231
A Howard M Sandler B Chen W Wang LC M Chen G Chu V Vasudevan Y Zhu R Pang Q Le H Adam 2019 Searching for mobileNetV3. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 1314–1324. Institute of Electrical and Electronics Engineers Inc
F Chollet 2017 Xception: Deep learning with depthwise separable convolutions. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017-Janua, 1800–1807. https://doi.org/10.1109/CVPR.2017.195
S Mehta M Rastegari 2022 Separable Self-attention for Mobile Vision Transformers.
W Yu P Zhou S Yan X Wang 2023 InceptionNeXt: When Inception Meets ConvNeXt.
Tu, Z., Talebi, H., Zhang, H., Yang, F., Milanfar, P., Bovik, A., Li, Y.: MaxViT: Multi-axis Vision Transformer. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 13684, 459–479 (2022). https://doi.org/10.1007/978-3-031-20053-3_27
Z Liu Y Lin Y Cao H Hu Y Wei Z Zhang S Lin B Guo 2021 Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF international conference on computer vision. pp. 10012–10022
H Touvron M Cord M Ai DeiT III: Revenge of the ViT. 1–27
H Touvron P Bojanowski M Caron M Cord A El-Nouby E Grave G Izacard A Joulin G Synnaeve J Verbeek H Jégou 2021 ResMLP: Feedforward networks for image classification with data-efficient training.
K Han Y Wang Q Tian J Guo C Xu C Xu 2019 GhostNet: More Features from Cheap Operations.
Y Tang K Han J Guo C Xu C Xu Y Wang 2022 GhostNetV2: Enhance Cheap Operation with Long-Range Attention.
PKA Vasu J Gabriel J Zhu O Tuzel A Ranjan 2023 FastViT: A Fast Hybrid Vision Transformer using Structural Reparameterization.
W Yu C Si P Zhou M Luo Y Zhou J Feng S Yan X Wang 2022 MetaFormer Baselines for Vision.
S Woo J Park J-Y Lee IS Kweon 2018 CBAM: Convolutional Block Attention Module.
Ozkaraca, O., Bagrıacık, O.İ, Guruler, H., Khan, F., Hussain, J., Khan, J., Laila, U.E.: Dense CNN architecture using brain MRI images. Life 13, 349 (2023). https://doi.org/10.3390/life13020349
Rahman, T., Islam, M.S.: MRI brain tumor detection and classification using parallel deep convolutional neural networks. Measurement: Sensors. 26, 100694 (2023). https://doi.org/10.1016/j.measen.2023.100694
Muezzinoglu, T., Baygin, N., Tuncer, I., Barua, P.D., Baygin, M., Dogan, S., Tuncer, T., Palmer, E.E., Cheong, K.H., Acharya, U.R.: PatchResNet: multiple patch division-based deep feature fusion framework for brain tumor classification using MRI Images. J. Digit. Imaging 36, 973–987 (2023). https://doi.org/10.1007/s10278-023-00789-x
Tabatabaei, S., Rezaee, K., Zhu, M.: Attention transformer mechanism and fusion-based deep learning architecture for MRI brain tumor classification system. Biomed. Signal Process. Control 86, 105119 (2023). https://doi.org/10.1016/j.bspc.2023.105119
Deepak, S., Ameer, P.M.: Brain tumor categorization from imbalanced MRI dataset using weighted loss and deep feature fusion. Neurocomputing 520, 94–102 (2023). https://doi.org/10.1016/j.neucom.2022.11.039
Zulfiqar, F., Ijaz Bajwa, U., Mehmood, Y.: Multi-class classification of brain tumor types from MR images using EfficientNets. Biomed. Signal Process. Control 84, 104777 (2023). https://doi.org/10.1016/j.bspc.2023.104777
Ghassemi, N., Shoeibi, A., Rouhani, M.: Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control 57, 101678 (2020). https://doi.org/10.1016/j.bspc.2019.101678
Mehnatkesh, H., Jalali, S.M.J., Khosravi, A., Nahavandi, S.: An intelligent driven deep residual learning framework for brain tumor classification using MRI images. Expert Syst. Appl. 213, 119087 (2023). https://doi.org/10.1016/j.eswa.2022.119087
Swati, Z.N.K., Zhao, Q., Kabir, M., Ali, F., Ali, Z., Ahmed, S., Lu, J.: Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 75, 34–46 (2019). https://doi.org/10.1016/j.compmedimag.2019.05.001
Sajjad, M., Khan, S., Muhammad, K., Wu, W., Ullah, A., Baik, S.W.: Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 30, 174–182 (2019). https://doi.org/10.1016/j.jocs.2018.12.003
Rehman, A., Naz, S., Razzak, M.I., Akram, F., Imran, M.: A deep learning-based framework for automatic brain tumors classification using transfer learning. Circuits Syst Signal Process. 39, 757–775 (2020). https://doi.org/10.1007/s00034-019-01246-3
Mzoughi, H., Njeh, I., Wali, A., Slima, M., Ben BenHamida, A., Mhiri, C., Mahfoudhe, K.: Ben: deep multi-scale 3d convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 33, 903 915 (2020). https://doi.org/10.1007/s10278-020-00347-9
Sharif, M.I., Khan, M.A., Alhussein, M., Aurangzeb, K., Raza, M.: A decision support system for multimodal brain tumor classification using deep learning. Complex and Intelligent Systems. 8, 3007–3020 (2022). https://doi.org/10.1007/s40747-021-00321-0
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). This work was supported by the grant provided by TÜSEB under the “2023-C1-YZ” call and Project No: “33934”. We would like to thank TÜSEB for their financial support and scientific contributions. Experimental computations were carried out on the computing units at Igdir University's Artificial Intelligence and Big Data Application and Research Center.
Author information
Authors and Affiliations
Contributions
Ishak Pacal: Conceptualization, Methodology, Software, Investigation, Data curation, Validation, Supervision, Writing – original draft, Writing – review & editing. Omer Celik: Conceptualization, Methodology, Data curation, Writing – review & editing. Bilal Bayram: Conceptualization, Methodology, Reviewing, Validation, Data curation, Writing – review & editing. Antonio Cunha: Methodology, Reviewing, Validation, Writing – review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
No ethics approval was required for this work as it did not involve human subjects, animals, or sensitive data that would necessitate ethical review.
Consent to participate
No formal consent to participate was required for this work as it did not involve interactions with human subjects or the collection of sensitive personal information.
Consent to publish
This study did not use individual person’s data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pacal, I., Celik, O., Bayram, B. et al. Enhancing EfficientNetv2 with global and efficient channel attention mechanisms for accurate MRI-Based brain tumor classification. Cluster Comput (2024). https://doi.org/10.1007/s10586-024-04532-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10586-024-04532-1