Abstract
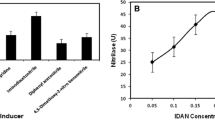
Nitrilase (EC 3.5.5.1) is an important biological catalyst with industrial application, which can directly convert nitrile compounds into corresponding carboxylic acids and ammonia under mild conditions. Nitrilase is used to convert molecules containing nitrile groups into carboxylic acid derivatives, and it is also used in many industries such as the textile industry. Studies should be conducted on the production of nitrilase, which is widely used in industry and has the potential to be used in other industrial areas, from different sources. Since reducing the cost in the industry will reflect positively on the consumer, high-activity nitrilases should be obtained. For this purpose, optimization of the production mediums gains importance in the production of nitrilase from microbial sources. The present study deals with the production and optimization of the microbial nitrilase (EC 3.5.5.1). Microbial strains stored in the laboratory were used in this study. All selected strains (Pseudomonas aeruginosa (ATCC 27853), Enterococcus faecalis (ATCC 29212), Pseudomonas putida (ATCC 17514), Pseudomonas fluorescens (DSM 6521), Pseudomonas oleovorans (DSM 50188), and Micrococcus luteus (DSM 20030)) from stock were screened on 3 different mediums. The acetonitrile showed to be an inducer for nitrilase production. The most active bacterium produced about 41.11 U/ml of nitrilase activity and was identified as Pseudomonas putida (ATCC 17514). Maximum nitrilase production was obtained using the medium (3) with 0.50 g/L (v/v) acetonitrile as an inducer. The effects of different parameters (chemical and physical) on the nitrilase (EC 3.5.5.1) production were studied. When bacterial growth at pH 8.0 and 28 ºC was cultured for 20 h, and medium containing starch as a carbon source and peptone-yeast extract as a nitrogen source, the nitrilase activity peaked at 172.76 U/ml. Enzyme activity could be increased 4.2 times with optimization studies. By optimization of enzyme production, a significant increase in activity may be achieved. In this study, when compared with similar studies about nitrilase production from bacteria in the literature, high-activity nitrilase production from Pseudomonas putida (ATCC 17514) was achieved.
Graphical Abstract







Similar content being viewed by others
References
Ramteke PW, Maurice NG, Joseph B, Wadher BJ (2013) Nitrile-converting enzymes: an eco-friendly tool for industrial biocatalysis. Biotechnol Appl Biochem 60:459–481
Dennett GV, Blamey JM (2016) A new thermophilic nitrilase from an antarctic hyperthermophilic microorganism. Front Bioeng Biotechnol 4:1–9
Toprak A, Tükel SS, Deniz Yildirim D (2021) Stabilization of multimeric nitrilase via different immobilization techniques for hydrolysis of acrylonitrile to acrylic acid. Biocatal Biotransfor 39:221–231
Ogunyemi AK, Buraimoh OM, Ogunyemi BC, Samuel TA, Ilori MO, Amund OO (2022) Nitrilase gene detection and nitrile metabolism in two bacterial strains associated with waste streams in Lagos, Nigeria. Bull Natl Res Cent 46:151–161
Banerjee A, Sharma R, Banerjee UC (2002) The nitrile-degrading enzymes: current status and future prospects. Appl Microbiol Biotechnol 60:33–44
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:142–159
Zhang X, Wang C, Ge Y, Meng Q, Zhang Y (2022) Constitutive secretory expression and characterization of nitrilase from Alcaligenes faecalis in Pichia pastoris for production of R-mandelic acid. Biotechnol Appl Biochem 69:587–595
Babu V, Shilpi CB (2010) Nitrile-metabolizing potential of Amycolatopsis sp. IITR215. Process Biochem 45:866–873
Zou SP, Huang JW, Xue YP, Zheng YG (2018) Highly efficient production of 1-cyanocyclohexaneacetic acid by cross-linked cell aggregates (CLCAs) of recombinant E. coli harboring nitrilase gene. Process Biochem 65:93–99
Xie ZY, Feng JL, Garcia E, Bernett M, Yazbeck D, Tao JH (2006) Cloning and optimization of a nitrilase for the synthesis of (3S)-3-cyano-5-methyl hexanoic acid. J Mol Catal B: Enzym 41:75–80
Cooling FB, Fager SK, Fallon RD, Folsom PW, Gallagher FG, Gavagan JE, Hann EC, Herkes FE, Phillips RL, Sigmund A, Wagner LW, Wu W, DiCosimo R (2001) Chemoenzymatic production of 1,5-dimethyl-2-piperidone. J Mol Catal B: Enzym 11:295–306
Shaw NM, Robins KT, Kiener A (2003) Lonza: 20 years of biotransformations. Adv Synth Catal 345:425–435
Liu ZQ, Zhang XH, Xue YP, Xu M, Zheng YG (2014) Improvement of Alcaligenes faecalis nitrilase by gene site saturation mutagenesis and its application in stereospecific biosynthesis of (R)-(−)-mandelic acid. J Agric Food Chem 62:4685–4694
Zhang XH, Liu ZQ, Xue YP, Wang YS, Yang B, Zheng YG (2017) Production of R-mandelic acid using nitrilase from recombinant E. coli cells immobilized with tris (hydroxymethyl) phosphine. Appl Biochem Biotechnol 184:1024–1035
Xue YP, Shi CC, Xu Z, Jiao B, Liu ZQ, Huang JF, Zheng YG, Shen YC (2015) Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering. Adv Synth Catal 357:1741–1750
Xue YP, Jiao B, Hua DE, Cheng F, Liu ZQ, Zheng YG (2017) Improving catalytic performance of an arylacetonitrilase by semirational engineering. Bioprocess Biosyst Eng 40:1565–1572
Panova A, Mersinger LJ, Liu Q, Foo T, Roe DC, Spillan WL, Sigmund AE, Ben-Bassat A, Wagner LW, O’Keefe DP, Wu S, Petrillo KL, Payne MS, Breske ST, Gallagher FG, DiCosimo R (2007) Chemoenzymatic synthesis of glycolic acid. Adv Synth Catal 349:1462–1474
Thuku RN, Brady D, Benedik MJ, Sewell BT (2009) Microbial nitrilases: versatile spiral forming, industrial enzymes. J Appl Microbiol 106:703–727
Gupta V, Gaind S, Verma PK, Sood N, Srivastava AK (2010) Purification and characterization of intracellular nitrilases from Rhodococcus sp.- potential role of periplasmic nitrilase. Afr J Microbiol Res 4:1148–1153
O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolysing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Vejvoda V, Kubáč D, Davidová A, Kaplan O, Šulc M, Šveda O, Martínková L (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Process Biochem 45:1115–1120
Petříčková A, Veselá AB, Kaplan O, Kubáč D, Uhnáková B, Malandra A, Felsberg J, Rinágelová A, Weyrauch P, Křen V, Bezouška K, Martínková L (2012) Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol 93:1553–1561
Sharma NN, Sharma M, Bhalla TC (2011) An improved nitrilase-mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol 38:1235–1243
Schreiner U, Hecher B, Obrowsky S, Waich K, Klempier N, Steinkellner G, Gruber K, Rozzell JD, Glieder A, Winkler M (2010) Directed evolution of Alcaligenes faecalis nitrilase. Enzyme Microb Technol 47:140–146
Martínková L, Vejvoda V, Kaplan O, Kubác D, Malandra A, Cantarella M (2009) Fungal nitrilases as biocatalysts: recent developments. Biotechnol Adv 27:661–670
Shen M, Liu ZQ, Zheng YG, Shen YC (2009) Enhancing Endo-nitrilase production by a newly isolated Arthrobacter nitroguajacolicus ZJUTB06-99 through optimization of culture medium. Biotechnol Bioproc Eng 14:795–802
Zhu D, Mukherjee C, Yang Y, Rios BE, Gallagher DT, Smith NN, Biehl ER, Hua L (2007) A new nitrilase from Bradyrhizobium japonicum USDA 110: gene cloning, biochemical characterization, and substrate specificity. J Biotechnol 133:327–333
Kaplan O, Vejvoda V, Plíhal O, Pompach P, Kavan D, Bojarová P, Bezouška K, Macková M, Cantarella M, Jirků V, Křen V, Ludmila Martínková L (2006) Purification and characterization of a nitrilase from Aspergillus niger K10. Appl Microbiol Biotechnol 73:567–575
Banerjee A, Kaul P, Banerjee UC (2006) Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol 184:407–418
Banerjee A, Kaul P, Banerjee UC (2006) Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol 72:77–87
Naik SC, Kaul P, Barse B, Banerjee A, Banerjee UC (2008) Studies on the production of enantioselective nitrilase in a stirred tank bioreactor by Pseudomonas putida MTCC 5110. Bioresour Technol 99:26–31
Fischer-Colbrie G, Matama T, Heumann S, Martinkova L, Paulo AC, Guebitz G (2007) Surface hydrolysis of polyacrylonitrile with nitrile hydrolysing enzymes from Micrococcus luteus BST20. J Biotechnol 129:62–68
Vikas V, Preeti S (2015) Soil as a Source for potential nitrilase producer. All Res J Biol 6:10–15
Shariati-Rad M, Qanei S (2022) Determination of ammonia based on experimental design and Berthelot reaction. J Environ Anal Chem. https://doi.org/10.1080/03067319.2022.2106429
Zhu X, Gong J, Li H, Lu Z, Zhou Z, Shi J, Xu Z (2014) Screening, identification and culture optimization of a newly isolated aromatic nitrilase-producing bacterium-Pseudomonas putida CGMCC3830. Chin J Biotechnol 30:412–424
Badoei-Dalfard A, Ramezani-Pour N, Karami Z (2016) Production and characterization of a nitrilase from Pseudomonas aeruginosa RZ44 and its potential for nitrile biotransformation. Iran J Biotechnol 14:142–153
Ramezani-Pour N, Badoei-Dalfard A, Namaki-Shoushtari A, Karami Z (2015) Nitrile-metabolizing potential of Bacillus cereus strain FA12; Nitrilase production, purification, and characterization. Biocatal Biotransfor 33:156–166
Komeda H, Harada H, Washika S, Sakamoto T, Ueda M, Asano Y (2004) A novel R-stereo selective amidase from Pseudomonas sp. MCI3434 acting on piperazine-2-tertbutyl carboxamide. Eur J Biochem 271:1580–1590
Kim JS, Tiwari MK, Moon HJ, Jeya M, Ramu T, Oh DK, Kim IW, Lee JK (2009) Identification and characterization of a novel nitrilase from Pseudomonas fluorescens Pf-5. Appl Microbiol Biotechnol 83:273–283
Kiziak C, Stolz A (2009) Identification of amino acid residues responsible for the enantioselectivity and amide formation capacity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Appl Environ Microbiol 75:5592–5599
Kumar S, Mohan U, Kamble AL, Pawar S, Banerjee UC (2010) Cross-linked enzyme aggregates of recombinant Pseudomonas putida nitrilase for enantioselective nitrile hydrolysis. Bioresour Technol 101:6856–6858
Agarwal A, Nigam VK (2014) Nitrilase mediated conversion of Indole-3-acetonitrile to Indole-3-acetic acid. Biocatal Agric Biotechnol 3:351–357
Makhongela HS, Glowacka AE, Agarkar VB, Sewell BT, Weber B, Cameron RA (2007) A novel thermostable nitrilase superfamily amidase from Geobacillus pallidus showing acyl transfer activity. Appl Microbiol Biotechnol 75:801–811
Chand D, Kumar H, Sankhian UD, Kumar D, Vitzthum F, Bhalla TC (2004) Treatment of simulated wastewater containing toxic amides by immobilized Rhodococcus rhodochrous NHB-2 using a highly compact 5-stage plug flow reactor. World J Microbiol Biotechnol 20:679–686
Reisinger Ch, Osprian I, Glieder A, Schoemaker HE, Griengl H, Schwab H (2004) Enzymatic hydrolysis of cyanohydrins with recombinant nitrile hydratase and amidase from Rhodococcus erythropolis. Biotechnol Lett 26:1675–1680
Ma Y, Yu HM, Pan W, Liu C, Zhang S, Shen Z (2010) Identification of nitrile hydratase-producing Rhodococcus ruber TH and characterization of an amiE-negative mutant. Bioresour Technol 101:285–291
Dong HP, Liu ZQ, ZhengYG ShenYC (2011) Medium optimization for nitrilase production by newly isolated Rhodococcus erythropolis ZJB-0910 using statistical designs. Chem Biochem Eng 25:351–358
He YC, Xu JH, Xu Y, Ouyang LM, Pan J (2007) Biocatalytic synthesis of (R)-(-)-mandelic acid from racemic mandelonitrile by a newly isolated nitrilase-producer Alcaligenes sp. ECU0401. Chin Chem Lett 18:677–680
He YC, Xu JH, Su JH, Zhou L (2010) Bioproduction of glycolic acid from glycolonitrile with a new bacterial isolate of Alcaligenes sp. ECU0401. Appl Biochem Biotechnol 160:1428–1440
Dinarvand M, Rezaee M, Masomian M, Jazayeri SD, Zareian M, Abbasi S, Ariff AB (2013) Effect of C/N ratio and media optimization through response surface methodology on simultaneous productions of intra- and extracellular inulinase and invertase from Aspergillus niger ATCC 20611. Biomed Res Int 2013:1–13
Klinfoong R, Thummakasorn C, Ungwiwatkul S, Boontanom P, Chantarasiri A (2022) Diversity and activity of amylase-producing bacteria isolated from mangrove soil in Thailand. Biodiversitas 23:5519–5531
Aiyer PVD (2004) Effect of C: N ratio on alpha amylase production by Bacillus licheniformis SPT 27. Afr J Biotechnol 3:519–522
Huang S, Huang D, Wu Q, Hou M, Tang X, Zhou J (2020) Effect of environmental C/N ratio on activities of lignin-degrading enzymes produced by Phanerochaete chrysosporium. Pedosphere 30:285–292
Nageshwar YVD, Sheelu G, Shambhu RR, Muluka H, Mehdi N, Malik MS, Kamal A (2010) Optimization of nitrilase production from Alcaligenes faecalis MTCC 10757 (IICT-A3): effect of inducers on substrate specificity. Bioprocess Biosyst Eng 34:515
Bisswanger H (2014) Enzyme assays Perspect Sci 1:41–55
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malaka, N.C., Akkaya, A. Optimization of Growth for Nitrilase Producing Bacteria. Catal Lett 154, 1232–1241 (2024). https://doi.org/10.1007/s10562-023-04377-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10562-023-04377-0