Abstract
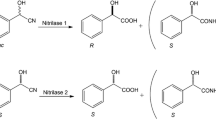
Nitrilases from Aspergillus niger CBS 513.88, A. niger K10, Gibberella moniliformis, Neurospora crassa OR74A, and Penicillium marneffei ATCC 18224 were expressed in Escherichia coli BL21-Gold (DE3) after IPTG induction. N. crassa nitrilase exhibited the highest yield of 69,000 U L−1 culture. Co-expression of chaperones (GroEL/ES in G. moniliformis and P. marneffei; GroEL/ES and trigger factor in N. crassa and A. niger CBS 513.88) enhanced the enzyme solubility. Specific activities of strains expressing the former two enzymes increased approximately fourfold upon co-expression of GroEL/ES. The enzyme from G. moniliformis (co-purified with GroEL) preferred benzonitrile as substrate (K m of 0.41 mM, V max of 9.7 μmol min−1 mg−1 protein). The P. marneffei enzyme (unstable in its purified state) exhibited the highest V max of 7.3 μmol min−1 mg−1 protein in cell-free extract, but also a high K m of 15.4 mM, for 4-cyanopyridine. The purified nitrilases from A. niger CBS 513.88 and N. crassa acted preferentially on phenylacetonitrile (K m of 3.4 and 2.0 mM, respectively; V max of 10.6 and 17.5 μmol min−1 mg−1 protein, respectively), and hydrolyzed also (R,S)-mandelonitrile with higher K m values. Significant amounts of amides were only formed by the G. moniliformis nitrilase from phenylacetonitrile and 4-cyanopyridine.


Similar content being viewed by others
References
Almatawah QA, Cramp R, Cowan DA (1999) Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles 3:283–291
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Banerjee A, Dubey S, Kaul P, Barse B, Piotrowski M, Banerjee UC (2009) Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol Biotechnol 41:35–41
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chao YP, Chiang CJ, Lo TE, Fu H (2000) Overproduction of D-hydantoinase and carbamoylase in a soluble form in Escherichia coli. Appl Microbiol Biotechnol 54:348–353
Chmura A, Shapovalova AA, van Pelt S, van Rantwijk F, Tourova TP, Muyzer G, Sorokin DYu (2008) Utilization of arylaliphatic nitriles by haloalkaliphilic Halomonas nitrilicus sp. nov. isolated from soda soils. Appl Microbiol Biotechnol 81:371–378
Fernandes BCM, Mateo C, Kiziak C, Chmura A, Wacker J, van Rantwijk F, Stolz A, Sheldon RA (2006) Nitrile hydratase activity of a recombinant nitrilase. Adv Synth Catal 348:2597–2603
Goldlust A, Bohak Z (1989) Induction, purification, and characterization of the nitrilase of Fusarium oxysporum f. sp. melonis. Biotechnol Appl Biochem 11:581–601
Gupta R, Lakshmipathy SK, Chang HC, Etchells SA, Hartl FU (2010) Trigger factor lacking the PPIase domain can enhance the folding of eukaryotic multi-domain proteins in Escherichia coli. FEBS Lett 584:3620–3624
Haacke A, Fendrich G, Ramage P, Geiser M (2009) Chaperone over-expression in Escherichia coli: apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expres Purif 64:185–193
Harper DB (1977) Fungal degradation of aromatic nitriles. Enzymology of C-N cleavage by Fusarium solani. Biochem J 167:685–692
Kaplan O, Vejvoda V, Plíhal O, Pompach P, Kavan D, Bojarová P, Bezouška K, Macková M, Cantarella M, Jirků V, Křen V, Martínková L (2006a) Purification and characterization of a nitrilase from Aspergillus niger K10. Appl Microbiol Biotechnol 73:567–575
Kaplan O, Nikolaou K, Pišvejcová A, Martínková L (2006b) Hydrolysis of nitriles and amides by filamentous fungi. Enzyme Microb Technol 38:260–264
Kaplan O, Bezouška K, Plíhal O, Ettrich R, Kulik N, Vaněk O, Kavan D, Benada O, Malandra A, Šveda O, Veselá AB, Rinágelová A, Slámová K, Cantarella M, Felsberg J, Dušková J, Dohnálek J, Kotik M, Křen V, Martínková L (2011a) Heterologous expression, purification and characterization of nitrilase from Aspergillus niger K10. BMC Biotechnol 11:2
Kaplan O, Bezouška K, Malandra A, Veselá AB, Petříčková A, Felsberg J, Rinágelová A, Křen V, Martínková L (2011b) Genome mining for the discovery of new nitrilases in filamentous fungi. Biotechnol Lett 33:309–312
Kato Y, Ooi R, Asano Y (2000) Distribution of aldoxime dehydratase in microorganisms. Appl Environ Microbiol 66:2290–2296
Kiziak C, Stolz A (2009) Identification of amino acid residues responsible for the enantioselectivity and amide formation capacity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Appl Environ Microbiol 75:5592–5599
Kiziak C, Conradt D, Stolz A, Mattes R, Klein J (2005) Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639–3648
Kiziak C, Klein J, Stolz A (2007) Influence of different carboxy-terminal mutations on the substrate-, reaction- and enantiospecificity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Protein Eng Des Sel 20:385–396
Laemmli UK (1970) Cleavage of the structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685
Layh N, Parratt J, Willetts A (1998) Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B-Enz 5:467–474
Lévy-Schil S, Soubriere F, Crutz-Le Coq AM, Faucher D, Crouzet J, Pétré D (1995) Aliphatic nitrilase from a soil-isolated Comamonas testosteroni sp.: gene cloning and overexpression, purification and primary structure. Gene 161:15–20
Lund PA (2009) Multiple chaperonins in bacteria—why so many? FEMS Microbiol Rev 33:785–800
Malandra A, Cantarella M, Kaplan O, Vejvoda V, Uhnáková B, Štěpánková B, Kubáč D, Martínková L (2009) Continuous hydrolysis of 4-cyanopyridine by nitrilases from Fusarium solani and Aspergillus niger K10. Appl Microbiol Biotechnol 85:277–284
Martínková L, Vejvoda V, Kaplan O, Kubáč D, Malandra A, Cantarella M, Bezouška K, Křen V (2009) Fungal nitrilases as biocatalysts: recent developments. Biotechnol Adv 27:661–670
Nishihara K, Kanemori M, Yanagi H, Yura T (2000) Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol 66:884–889
O'Reilly C, Turner PD (2003) The nitrilase family of CN hydrolysing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Osswald S, Wajant H, Effenberger F (2002) Characterization and synthetic applications of recombinant AtNIT1 from Arabidopsis thaliana. Eur J Biochem 269:680–687
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Rustler S, Motejadded H, Altenbuchner J, Stolz A (2008) Simultaneous expression of an arylacetonitrilase from Pseudomonas fluorescens and a (S)-oxynitrilase from Manihot esculenta in Pichia pastoris for the synthesis of (S)-mandelic acid. Appl Microbiol Biotechnol 80:87–97
Sabate R, de Groot NS, Ventura S (2010) Protein folding and aggregation in bacteria. Cell Mol Life Sci 67:2695–2715
Sareen D, Sharma R, Vohra RM (2001) Chaperone-assisted overexpression of an active D-carbamoylase from Agrobacterium tumefaciens AM 10. Protein Expres Purif 23:374–379
Sosedov O, Matzer K, Bürger S, Kiziak C, Baum S, Altenbuchner J, Chmura A, van Rantwijk F, Stolz A (2009) Construction of recombinant Escherichia coli catalysts which simultaneously express an (S)-oxynitrilase and different nitrilase variants for the synthesis of (S)-mandelic acid and (S)-mandelic amide from benzaldehyde and cyanide. Adv Synth Catal 351:1531–1538
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thuku RN, Weber BW, Varsani A, Sewell BT (2007) Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J 274:2099–2108
Thuku RN, Brady D, Benedik MJ, Sewell BT (2009) Microbial nitrilases: versatile, spiral forming, industrial enzymes. J Appl Microbiol 106:703–727
Vejvoda V, Kaplan O, Bezouška K, Pompach P, Šulc M, Cantarella M, Benada O, Uhnáková B, Rinágelová A, Lutz-Wahl S, Fischer L, Křen V, Martínková L (2008) Purification and characterization of a nitrilase from Fusarium solani O1. J Mol Catal B-Enz 50:99–106
Vejvoda V, Kubáč D, Davidová A, Kaplan O, Šulc M, Šveda O, Chaloupková R, Martínková L (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Proc Biochem 45:1115–1120
Wu SJ, Fogiel AJ, Petrillo KL, Hann EC, Mersinger LJ, DiCosimo R, O'Keefe DP, Ben-Bassat A, Payne MS (2007) Protein engineering of Acidovorax facilis 72W nitrilase for bioprocess development. Biotechnol Bioeng 97:689–693
Wu SJ, Fogiel AJ, Petrillo KL, Jackson RE, Parker KN, DiCosimo R, Ben-Bassat A, O'Keefe DP, Payne MS (2008) Protein engineering of nitrilase for chemoenzymatic production of glycolic acid. Biotechnol Bioeng 99:717–720
Acknowledgments
The authors wish to thank Hynek Mrazek MSc. for his technical help with nitrilase purification. Financial support via projects P504/11/0394, 305/09/H008 (Czech Science Foundation), IAA500200708 (Grant Agency of the Academy of Sciences of the Czech Republic), LC06010, OC09046 (Ministry of Education of the Czech Republic), COST/ESF CM0701 (STSM fellowships COST-STSM-CM0701-4765 and −4766 to A. Malandra) and Institutional Research Concept AV0Z50200510 (Institute of Microbiology) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Alena Petříčková and Alicja B. Veselá contributed equally to this work.
An erratum to this article is available at http://dx.doi.org/10.1007/s00253-013-5204-3.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Nucleotide sequences of the nitrilase genes from P. marneffei ATCC18224 (GenBank JN012233) and A. niger K10 (GenBank JN243351) adapted for expression in E. coli using GeneArt (Regensburg, Germany) software. (DOC 29 kb)
Fig. S2
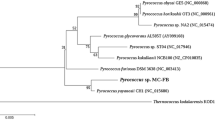
Phylogenetic tree of fungal nitrilase/cyanide hydratase enzymes. The search was performed with the sequence of nitrilase from A. niger K10 (Kaplan et al. 2011a; marked with asterisks) as the query using BLASTP. Nitrilases expressed and characterized by us are highlighted (DOC 693 kb)
Fig. S3
Multiple sequence alignment using ClustalW software (Thompson et al. 1994) of nitrilases from Gibberella moniliformis (GiMon Nit; GenBank ABF83489), Neurospora crassa OR74A (NeCra Nit; GenBank CAD70472), Aspergillus niger K10 (AsNig Nit1; GenBank ABX75546; Kaplan et al. 2011a), Penicillium marneffei ATCC18224 (PeMar Nit; GenBank XP_002144951), and Aspergillus niger CBS 513.88 (AsNig Nit2; GenBank XP_001397369). Clustal co clustal consensus, asterisk identical aa residues, colon similar aa residues, dot less similar aa residues (DOC 36 kb)
Table S4
Screening of chaperone effects on nitrilase activity in E. coli BL21-Gold(DE3) strains (DOC 68.5 kb)
Rights and permissions
About this article
Cite this article
Petříčková, A., Veselá, A.B., Kaplan, O. et al. Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol 93, 1553–1561 (2012). https://doi.org/10.1007/s00253-011-3525-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3525-7