Abstract
Purpose
How to factor both tumor burden and oncogenic genomic mutations as variables to predict the outcome of endocrine-based therapy (ET) in ER-positive/HER2-negative metastatic breast cancer patients (MBC) remains to be explored.
Method
Blood samples prospectively collected from 163 ER-positive/HER2-negative female MBC patients, before ET, were used for cell-free tumor DNA (cfDNA) analysis. cfDNA was subjected to next-generation sequencing (NGS) to interrogate oncogenic PIK3CA hotspot and TP53 DNA-binding domain (DBD) mutations, including single nucleotide variants (SNVs) or small insertions and deletions (InDels). The variant calling threshold was set at 0.5%. Progression-free survival (PFS) was measured from the start of the ET treatment to the time of disease progression of the same treatment regimen.
Results
Overall, the median PFS was 8.3 months (95% CI 5.7–11.1 months). The median cfDNA was 38.5 ng (range 4.4–1935 ng). The proportion of patients with PIK3CA and TP53 alterations were 25.1 and 15.3%, respectively. Patients with high total cfDNA (HR 1.74, p = 0.003), PIK3CA mutation (HR 1.74, p = 0.007), and TP53 mutation (HR 1.64, p = 0.047) in liquid biopsy conferred worse outcome after ET. Even for patients with low tumor burden, the detrimental effect of PIK3CA or TP53 mutation remained significant (p < 0.001). For patients with either PIK3CA (p < 0.001) or TP53 mutation (p = 0.004), there was significant positive correlation between allele frequency (AF) and total cfDNA.
Conclusion
After adjustment of cfDNA level, PIK3CA and TP53 mutations observed in liquid biopsy exerted detrimental effects on the outcome of ET-based regimens. The AF of PIK3CA or TP53 may be a surrogate marker for PFS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite the progress in the endocrine therapy (ET)-based regimens of endocrine receptor (ER)-positive human epidermal growth factor receptor (HER2)-negative metastatic breast cancer (MBC), the prognosis and the survival outcome of different treatment regimens vary far and wide. A better estimate of treatment outcomes could facilitate communication of physicians and patients and the selection of the optimal treatment for these patients.
Tumor burden plays a critical impact on prognosis but it may not be easily quantified and estimated for each patient. For ER-positive MBC patients, estimating tumor burden may be further complicated by the common involvement of bone metastases that is difficult to measure the extent and size of metastatic tumors. The genomic DNA of cancer cells could shed into the bloodstream in the form of cell-free tumor DNA (cfDNA) and is characterized by shorter fragmented reads, ranging from 100 to 200 base pairs [1]. Studies in many cancer types have shown that the total amount of cfDNA is a reflection of total tumor burden and associated with prognosis [1,2,3,4,5,6]. In addition, these cfDNA could also be used as sources for identification of of specific genetic alterations that are also important in the evaluation of prognosis [7,8,9].
PIK3CA and TP53 are two genes that have been shown to affect the treatment outcome and prognosis of MBC. As PIK3CA activation via mutation is one the resistance mechanisms of chemotherapy and anti-HER2 therapy [10, 11], some studies have shown that a ET-based combination regimen may still provide similar proportion of benefit to both PIK3CA wild type and mutant patients [12, 13]. However, because these studies may incorporate patients with a wide range of tumor burden, parsing out the factor of tumor burden may provide insights to our understanding of the clinical impact of PIK3CA mutation. The tumor suppressor TP53 is one of the most common mutated gene in cancer but it’s role in MBC is less investigated. Although TP53 alterations are more common in triple-negative breast cancer [6], TP53 had strong negative prognostic impact when present in ER-positive early breast cancer [14]. Despite studies have suggested that alterations of oncogenic mutations TP53 are associated with worse outcome [15], few incorporated or adjusted the factor of tumor burden in their studies.
Liquid biopsy of a patient’s blood could provide information regarding the total cfDNA as well as tumor-specific genomic alterations via next-generation sequencing (NGS) [1, 2]. Albeit there are multiple research- or commercial-based cfDNA targeted panel of liquid biopsy in the clinical field, few integrated tumor burden and oncogenic mutations of breast cancers into the profiling and its association with treatment outcome.
In this study, we aim to prospectively collect ET-based treatment outcome of ER-positive HER2-negative MBC patients and the clinical utility of liquid biopsy cfDNA testing. Instead of collecting a broad mutation profile, we designed a targeted amplicon sequencing panel focused on oncogenic mutations of the two most common mutations in ER-positive breast cancer, namely PIK3CA and TP53 and incorporated the total cfDNA as one of the reporting endpoints. Our study provides evidence that a focused panel of genes and inclusion of total cfDNA is adequate to provide insights of the impact on ET-based treatment for ER-positive HER2-negative MBC patients.
Patients and methods
Patient enrollment
ER + /HER2- MBC patients were eligible if they were older than age 20 and were intended for ET-based regimens. ET included single agent endocrine therapy agent or in combination with targeted agents such as cycline-dependent kinase (CDK) 4/6 inhibitor or everolimus (a mammalian target of rapamycin (mTOR) inhibitor). Patients who received endocrine therapy with metronomic chemotherapy were also eligible. The determination of ER and HER2 status was according to the ASCO guideline as previously reported [16, 17]. Repeated biopsy after metastatic diagnosis was not required and the status of ER and HER2 was based on the most recent pathological report (primary or metastatic pathological reports were both eligible). Subgroup analysis for the ET plus everolimus subgroup was pre-specified in the protocol. This study was approved by the Research Ethics Committee of National Taiwan University Hospital (201411008RIND & 201703138RIPD) and Institutional Review Board of Tri-Service General Hospital (1-107-05-003).
DNA extraction and NGS
Blood sample was collected in the Cell-Free DNA Blood Collection Tubes (BCT, Streck, La Vista, NE) [18] followed by a two-step centrifugation (1600×g then 17000×g, both for 10 min at room temperature) separation procedure. After separation, 2.5–4 ml of plasma was used to extract cfDNA with the QIAamp circulating nucleic acid kit according to the manufacturer’s protocol (Qiagen). DNA concentration and integrity were determined using the Quant-iT dsDNA HS Assay (Invitrogen) and the Fragment Analyzer (Advanced Analytical Technologies, Inc.), respectively. A total of 20 ng of cfDNA would be was subsequently PCR-amplified for 26 cycles with a multiplex panel consisting of primer pairs covering regions of TP53 DNA-binding domain (DBD) and hotspot PIK3CA mutations, which included a total of 13 amplicons containing 1137 base pairs. PCR products were ligated to barcode adapters and underwent further amplification, followed by emulsion PCR on the OneTouch System (Applied Biosystems). A maximum of 96 samples were pooled on the Ion PI™ Chip Kit on Ion Proton System with the aim for an average of 10,000 × average read depth. Twelve healthy volunteer’s PBMC were used as control.
Variant analysis
The analyzed variants included single nucleotide variants and small insertions and deletions with the limitation of variant detection set at 0.5%. The human genome sequence hg19 was used as the reference genome, and alignment and base calling were performed with the Torrent Suite Server version 4.4. Annotated plasma variants had to have at least 20 reads and the allelic fraction had to be above a background threshold of 7 Z-Scores from the mean of healthy donors. When developing this NGS assay and analysis, we assumed each sequencing position will be covered 1000 × reads. Due to the limit of detection being set to 0.5%, the variant read counts need to be larger than 20 read counts for further manual check. Annotated tumor variants had to have a variant frequency of at least 0.5%, or to be known hotspot variants identified as true variants after raw data analysis. For annotation, COSMIC (v74), dbSNP 138 and 1000 Genomes of the global population data (phase1) were used.
Statistical analysis
Progression-free survival was defined as the date of start of the treatment to the date of evidence of disease progression. Log-rank test and Cox proportional hazard methods were applied for the testing of genomic alteration and PFS outcome. Linear regression and Pearson correlation coefficients were used to examine the correlation between the allele frequency (AF) of cfDNA genomic alterations and PFS outcome. All statistical tests were performed using R 3.6.3.
Results
From Aug 2015 to May 2020, a total of 163 ER-positive HER2-negative MBC patients were prospectively enrolled into the study. All patients were women. The median age was 60 (range 32–92). The lines of treatment that patients intended to receive following the enrollment was 20, 34, 31, 15% for first-line, second-line, third-line, and fourth or later-lines, respectively. All patients received at least an endocrine therapeutic agent as part of the regimen. Thirteen percent of patients received ET as single agent, 48% received ET plus everolimus, 14% received ET plus a CDK4/6 inhibitor, and 17% of patients received ET plus metronomic chemotherapy. Fourteen (8.5%) patients received fulvestrant as part of the ET regimen. The detail treatment regimens are listed in Table 1.
The mutation profiles in ER-positive HER2-negative MBC patients
Using a 0.5% cut-off threshold for variant calling (see Patients and Methods), 57 (34.9%) patients had at least one PIK3CA or TP53 mutations detected in the plasma with 41 (25.1%) and 25 (15.3%) patients harbored PIK3CA and TP53 mutations, respectively. The proportions of patients with at least one genomic alteration did not differ significantly between lines of ET treatment (p = 0.81 for PIK3CA, p = 0.42 for TP53).
All patients with detectable PIK3CA cfDNA had single-point mutation and no small indels were detected. Three (8%) patients had 2 different PIK3CA mutation genotypes detected by liquid biopsy whereas all other patients had only one specific PIKCA mutation. The most common PI3KCA mutation hotspot was found at H1047 (53% of all PIK3CA alterations, including H1047R, H1047L and H1047Y). All identified PIK3CA mutations by liquid biopsy are shown in Fig. 1a. As for TP53, point mutations, frameshift changes and deletions (Fig. 1b) were identified. Owing to amplicon design limitations, all cfDNA TP53 mutations were located within the DNA-binding domain.
The effects of tumor burden on treatment outcome
Inclusive of all patients, the median PFS was 8.3 months (95% CI 5.7–11.1 months). The duration of PFS of each individual ET-based regimen and each line of ET-based regimens are shown in Table 1.
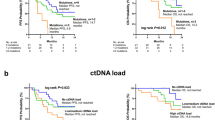
In here we relate tumor burden not only to the sheer volume of the disease but also to the aggressiveness of the tumor, as in contrast to some patients may have a moderate to high volume of tumor mass but with an indolent disease pattern. As cfDNA in the circulation is largely made up by genomic DNA fragments released from apoptotic or necrotic cancerous cells as a result of rapidly proliferation, the amount of cfDNA recovered can be used as surrogate for tumor burden evaluation [19, 20]. We also applied a cfDNA-tailored blood collection tube and algorithm to minimize the bias from normal cell circulating DNA [18]. We then explored the effect of tumor burden, as represented by total cfDNA, and oncogenic mutations PIK3CA and TP53 on the outcome of ET-based regimens. The median amount of recovered cfDNA in the cohort was 38.5 ng (range 4.4–1935 ng) (The total cfDNA histogram is shown in Supplementary Fig. 1). Using median of 38.5 as the threshold for high tumor burden, we found that patients with higher tumor burden was significantly associated with a worse outcome after ET-based regimens (median PFS 5.6 vs 13.3 months, log-rank p = 0.003, HR 1.74, 95% CI 1.12–2.51,) (Fig. 2a).
The effects of PIK3CA and TP53 oncogenic mutations on treatment outcome
Patients with PIK3CA mutation had worse PFS as compared to those without (median PFS 4.5 vs 10.3 month, log-rank p = 0.007, HR 1.74, 95% CI 1.16–2.62, Fig. 2b). We also compared the PFS results between patients with kinase domain (H1047X) mutation and other non-kinase domain PIK3CA mutations and found no significant differences (H1047X vs others median PFS 4.8 vs 5.8 months, p = 0.9). Moreover, plasma TP53 mutations also exerted an inferior outcome (median PFS 4.1 vs 8.9, HR 1.64, 95% CI 1.01–2.67, p = 0.044, Fig. 2c). Changes of TP53 protein were classified as missense or truncated based on predicted amino acid changes of the mutations and we did not find significant differences between the PFS results of these two groups (missense vs truncated TP53 5.6 vs 4.0 months, p = 0.23). The detrimental impact of TP53 on PFS remained significant in PIK3CA wild type patients (HR 3.28, 95% CI 1.44–7.48, p = 0.0048). We also examined the differences in median PFS between double WT, PIK3CA mutant only, TP53 mutant only, and patients with both PIK3CA and TP53 mutations. The PFS between the three groups with any mutations were not significantly different (Supplementary Fig. 2).
Correlation between tumor burden and PIK3CA/TP53 mutations
Because liquid biopsy by NGS is able to provide the allele frequency (AF) of each mutated variant, we aimed to understand if the AF of PIK3CA and TP53 oncogenic mutations could be representative of total tumor burden. Thus, the correlation between total cfDNA (in ng) and AF (in %) of either PIK3CA or TP53 mutation variants were examined. Patients with wild type PIK3CA or TP53 were considered to have 0% AF. Inclusive of all patients, regardless of the status of the oncogenic mutations, the correlation between total cfDNA and PIK3CA and TP53 AF was weak (Pearson correlation coefficient R = 0.34, p < 0.001) and non-existent (R = 0.055, p = 0.48), respectively (Fig. 3a, c). If the correlation analysis included only patients with PIK3CA or TP53 mutations (AF > 0) identified by liquid biopsy, the correlation coefficient increased to 0.53 (p < 0.001, Fig. 3b) and 0.43 (p = 0.033, Fig. 3d) for PIK3CA and TP53, respectively. When the correlation analysis was limited to patients with hotspot PIK3CA mutations (H1047R, E545K, E542K, and H1047L, comprising 77% of PIK3CA mutations in our cohort, Fig. 1a), PIK3CA AF and cfDNA remained positively correlated (R = 0.38, p = 0.018). Thus, PIK3CA mutation AF in the liquid biopsy had a stronger correlation with tumor burden.
Separating the effects of tumor burden and oncogenic mutations
Based on the mutation status of PIK3CA and TP53 mutation and the tumor burden, and we separated the patients into four groups (high vs low tumor burden and presence or no presence of cfDNA mutation). This was intended to understand the differential effect of these two factors on the treatment outcome of ET-based regimens. Not unexpectedly, patients with higher tumor burden and a presence of either PIK3CA or TP53 oncogenic mutation had the worse outcome (p < 0.001) (Fig. 4a). Interestingly, patients with a presence of PIK3CA or TP53 mutation but low tumor burden had similar PFS outcome with those with a high tumor burden but with no PIK3CA or TP53 oncogenic mutations (median PFS 5.8 vs 5.6 months), suggesting that the oncogenic mutations of PIK3CA and/or TP53 exerted additional resistance to the treatment of ET-base regimens in addition to the tumor burden status that was associated with worse PFS. These detrimental impact of PIK3CA or TP53 on PFS were confirmed after adjusting for the lines of treatment and the type of treatment (HR 1.89 95% CI 1.29–2.79, p = 0.001). The HR for patients with either a PIK3CA or TP53 mutation receiving ET-based regimen as first, second, third and fourth line were 2.09, 2.15, 2.45, and 1.13, respectively. The p-value for interaction was 0.54.
In the preplanned subgroup analysis of patients received ET + everolimus (n = 78), the detrimental effect of high tumor burden (HR 1.70, 95% CI 1.04–2.78, p = 0.03) and PIK3CA and/or TP53 oncogenic mutation (HR 1.67, 95% CI 1.02–2.76, p = 0.04) remained significant (Fig. 4b).
Discussion
In addition to the less-invasive nature, liquid biopsies contain genetic alterations that reflect intra- and inter-tumoral heterogeneity, which is valuable for direct personalized therapies. In our prospective study of 163 patients, we showed that total cfDNA and the presence of either PIK3CA or TP53 oncogenic mutation from liquid biopsy before the start of endocrine-based therapy were associated with worse PFS in ER + /HER2- MBC patients. In addition, after adjustment of other clinical variables such as the lines and the type of treatment, these two factors remained significantly associated with PFS. The prospective nature of the study and that we limited the recruitment to patients treated with ET-based regimens in a more homogenous population further lends strength to our findings.
While studies have reported the association between a worse outcome and the presence of PIK3CA or TP53 mutation, few included the tumor burden as a variable to adjust the impact of these oncogenic mutations. The tumor burden of ER-positive MBC may be especially hard to estimate because of the propensity for bone metastases. Although other factors such as peripheral blood mononuclear cell lysis or normal tissue shedding that could confound the interpretation of cfDNA, we specifically selected shorter fragments of cell-free DNA that are associated with tumor DNA [18]. Our findings that the amount of total cfDNA were inversely associated with PFS supports that total cfDNA may be considered as a surrogate marker of tumor burden in ER-positive MBC patients. However, we need to be cautious to use total cfDNA solely as a surrogate marker to assess tumor burden, which, while important, is complicated and has no standard procedures based on liquid biopsy results at present. Factors that could influence the interpretation of cfDNA such as tumor DNA shedding and the mutation fraction should also be considered before implantation of cfDNA as the sole biomarker for tumor burden.
Few prospective studies have accounted for the tumor burden when assessing the impact of PIK3CA and TP53 mutations on ET-based treatment based on liquid biopsy results. In this study, the inverse association of PIK3CA/TP53 mutations with PFS was evident in patients with high and low tumor burdens. Previous studies showed that PIK3CA mutation rate did not differ significantly between primary tumors and corresponding metastases, but the prognostic effect was stage-dependent. In early stage breast cancer patients, the presence of PIK3CA mutation was associated with a better disease-free survival and overall survival [21]. But in ER-positive metastatic breast cancer, the presence of PIK3CA mutation exerted a negative effect on survival [11]. The transition from favorable to the inferior impact of PIK3CA mutations supported our findings that PIK3CA had a significant negative survival impact at the early stage of metastatic disease and conferred resistance to treatments.
The findings that PIK3CA mutation in the cfDNA is a prognostic marker for worse PFS in ET-based regimens have multiple clinical implications. A recent randomized phase III study, SOLAR-1, demonstrated the superior efficacy of the combination of fulvestrant and alpelisib, a PI3k-alpha specific inhibitor, as compared to fulvestrant alone in ER-positive, HER2-negaitve MBC [22]. The benefit of alpelisib may even be stronger in patients whose PIK3CA were detected by cfDNA [23]. However, the benefit of alpelisib seemed to be more modest when used in a later line setting [24]. Incorporating our findings that PIK3CA mutation portends a worse prognosis even when tumor burden is low, patients should consider specific PIK3CA inhibitor in the earliest time after failure of standard first-line treatment of ET plus a CDK4/6 inhibitor.
Liquid biopsy may provide the variant AF of multiple genetic mutations in a single test but debate remains as to the variant AF of which gene may best represent tumor burden. In our study, the two genes selected for NGS are both common mutations of breast cancer. However, PIK3CA is an oncogene where hotspots are mostly single nucleotide variant and TP53 is a tumor suppressor gene that various types of genetic alterations could lead to a dysfunctional p53 protein [15, 25]. Our results suggested that in patients with PIK3CA mutation identified by liquid biopsy, the variant AF of PIK3CA strongly correlated with tumor burden. However, the correlation between TP53 variant AF and total cfDNA was less pronounced and the reason may be that amplicon-based NGS is unable to detect larger deletions, loss of heterozygosity, and structural variations. Thus, when an amplicon-based NGS panel is used for liquid biopsy testing, the variant AF of a oncogene mutation may provide a better reflection of tumor burden than tumor suppressor genes. However, the correlation between TP53 variant AF and total cfDNA was less pronounced, and the reason may be that amplicon-based NGS is unable to detect larger deletions, loss of heterozygosity, and structural variations. Thus, when an amplicon-based NGS panel is used for liquid biopsy testing, based on our study results, we propose that the variant AF of a bona fide oncogene mutation may better reflect tumor burden than tumor suppressor genes. Newer NGS technology that could detect a wider range of genomic alterations and structural variations associated with tumor suppressor genes in liquid biopsy could be more precise in using AF to reflect tumor suppressor gene mutated clones. Investigators could further validate this hypothesis and determine how to optimally use AF of various gene alterations in this era that dozens of genetic alterations are within reach in liquid biopsy samples. Furthermore, the dynamic changes of the AF of ctDNA in liquid biopsy could also reflect the changes of tumor heterogeneity (intra- and inter-tumor) and may reveal the potential resistance mechanism to ET-based regimens.
The success of BOLERO-2 study confirmed the role of everolimus in the treatment of ER-positive HER2-negative MBC [26]. Retrospective analysis of archival tumor specimens and droplet-digital PCR-based liquid biopsy from BOLERO-2 showed that the presence of PIK3CA mutation was a prognostic but not predictive biomarker [12, 27]. Although TP53 mutation status was examined, there was no formal report regarding the effect of TP53 on the treatment efficacy of exemestane plus everolimus in BOLERO-2 [12, 27]. Through NGS-based liquid biopsy, we prospectively confirmed that PIK3CA and TP53 mutations conferred a detrimental effect with shorter PFS in patients treated with the combination of ET + everolimus.
Our study has some caveats. It is widely accepted that the combination of an ET plus CDK4/6 inhibitor is the first-line treatment standard for ER-positive/HER2-negative MBC, but only 14% of the study population received first-line CDK4/6 inhibitors. Nested studies from phase III CDK4/6 inhibitor clinical trials have suggested that the presence of PIK3CA mutation did not have significant detrimental impact on PFS. [13, 28]. Thus, our conclusion that PIK3CA mutation is a marker for worse outcome for ET-based regimens in ER-positive MBC should be cautiously interpreted in the first-line setting with ET and CDK4/6 inhibitor combinations. Secondly, our analysis did not cover the entire coding regions of genes selected, which may account for the lower than expected mutation rate of TP53. In addition, large-segment deletions or gene rearrangements analysis is not allowed due to the limited length of cfDNA. However, because more than 95% of PIK3CA mutations in BC are hotspot mutations [29, 30], we are confident that our study design could pick up most of the PIK3CA mutations. Lastly, the amount of cfDNA could be influenced by the total amount of plasma extracted from patients. Although we did not record each sample’s exact whole blood and subsequent plasma volume, the protocol-based standardized procedures of blood draw, plasma separation, and cfDNA extraction made us believe that the error (bias) would more likely to be randomly distributed and have limited impact to the results.
In conclusion, we demonstrated plasma PIK3CA and TP53 mutations are independent predictive markers for shortened PFS in ER-positive BC receiving ET-based regimens. Besides, the level of plasma cfDNA and PIK3CA/TP53 AF could be a surrogate for tumor burden to correlate treatment outcomes. Liquid biopsy in ER-positive/HER2-negative MBC patients is achievable, provides prognosis information, and may also lead to different selection of treatment of the patients.
Data availability
The datasets generated and analysed during the current study are not publicly available becuase it was not permitted by our Research Ethics Committte. but may be available from the corresponding author on reasonable request.
References
Ignatiadis M, Sledge GW, Jeffrey SS (2021) Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol 18:297–312
Keller L, Belloum Y, Wikman H, Pantel K (2021) Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 124:345–358
Hemming ML, Klega KS, Rhoades J et al (2019) Detection of circulating tumor DNA in patients with leiomyosarcoma with progressive disease. JCO Precis Oncol 3:1–11
Shibayama T, Low SK, Ono M et al (2020) Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res Treat 180:331–341
Fernandez-Garcia D, Hills A, Page K et al (2019) Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res 21:149
Rossi G, Mu Z, Rademaker AW et al (2018) Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res 24:560–568
Jacot W, Dalenc F, Lopez-Crapez E et al (2019) PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res Treat 177:659–667
Keup C, Benyaa K, Hauch S et al (2020) Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell Mol Life Sci 77:497–509
Phallen J, Sausen M, Adleff V et al (2017) Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aan2415
Baselga J, Cortés J, Im SA et al (2014) Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol 32:3753–3761
Mosele F, Stefanovska B, Lusque A et al (2020) Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol 31:377–386
Hortobagyi GN, Chen D, Piccart M et al (2016) Correlative analysis of genetic alterations and everolimus benefit in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from BOLERO-2. J Clin Oncol 34:419–426
Tolaney SM, Toi M, Neven P et al (2022) Clinical significance of PIK3CA and ESR1 mutations in circulating tumor DNA: analysis from the MONARCH 2 study of abemaciclib plus fulvestrant. Clin Cancer Res 28:1500–1506
Lin CH, Chen IC, Huang CS et al (2015) TP53 mutational analysis enhances the prognostic accuracy of IHC4 and PAM50 assays. Sci Rep 5:17879
Razavi P, Chang MT, Xu G et al (2018) The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34(427–438):e426
Allison KH, Hammond MEH, Dowsett M et al (2020) Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J Clin Oncol 38:1346–1366
Wolff AC, Hammond MEH, Allison KH et al (2018) Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline focused update. J Clin Oncol 36:2105–2122
Medina Diaz I, Nocon A, Mehnert DH et al (2016) Performance of streck cfDNA blood collection tubes for liquid biopsy testing. PLoS ONE 11:e0166354
Bredno J, Lipson J, Venn O et al (2021) Clinical correlates of circulating cell-free DNA tumor fraction. PLoS ONE 16:e0256436
Alix-Panabieres C, Pantel K (2016) Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6:479–491
Zardavas D, Te Marvelde L, Milne RL et al (2018) Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol 36:981–990
Andre F, Ciruelos E, Rubovszky G et al (2019) Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 380:1929–1940
Ciruelos EM, Loibl S, Mayer IA et al (2021) Abstract PD2-06: clinical outcomes of alpelisib plus fulvestrant in hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer with PIK3CA alterations detected in plasma ctDNA by next-generation sequencing—biomarker analysis from the SOLAR-1 study. Cancer Research 81:206
Rugo HS, Lerebours F, Ciruelos E et al (2021) Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol 22:489–498
Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Baselga J, Campone M, Piccart M et al (2012) Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 366:520–529
Moynahan ME, Chen D, He W et al (2017) Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(-) advanced breast cancer: results from BOLERO-2. Br J Cancer 116:726–730
Hortobagyi GN, Stemmer SM, Burris HA et al (2018) Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann Oncol 29:1541–1547
Barone I, Brusco L, Fuqua SA (2010) Estrogen receptor mutations and changes in downstream gene expression and signaling. Clin Cancer Res 16:2702–2708
Dogruluk T, Tsang YH, Espitia M et al (2015) Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res 75:5341–5354
Funding
This study received grant support from Taiwan Ministry of Health and Welfare (MOHW107-TDU-B-211-114017, MOHW108-TDU-B-211-124017, MOHW109-TDU-B-211-134017, MOHW110-TDU-B-211-144017) and Novartis.
Author information
Authors and Affiliations
Contributions
Study conception: TWC, WH, KHT, YSL; Data Collection: all authors, Data analysis: TWC, WH, KHT, YSL; Manuscript preparation and confirmation: all authors.
Corresponding authors
Ethics declarations
Conflict of interest
WH, KWW, and KHT were employees of ACT Genomics Inc. TWC, MSD, CHL, DYC, ICC, ALC, and YSL have received honorarium from Novartis. YSL had received research funding from Novartis.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2023_6967_MOESM1_ESM.tiff
Supplementary file1 (TIFF 7607 KB)—Fig. 1 The distribution of extracted cell-free DNA (cfDNA). The dashed line indicates the median

10549_2023_6967_MOESM2_ESM.jpg
Supplementary file2 (JPG 1557 KB)—Fig. 2 The Kaplan-Meier curve of median progression-free survival (PFS) of patients based on the mutation status of PIK3CA and TP53. MT mutation, WT wild type
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, T.WW., Hsiao, W., Dai, MS. et al. Plasma cell-free tumor DNA, PIK3CA and TP53 mutations predicted inferior endocrine-based treatment outcome in endocrine receptor-positive metastatic breast cancer. Breast Cancer Res Treat 201, 377–385 (2023). https://doi.org/10.1007/s10549-023-06967-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06967-3