Abstract
Purpose
In this study, we aim to investigate the mutation spectrum of circulating tumor DNA among hormone receptor-positive metastatic breast cancer (HR-MBC) patients using ultradeep targeted resequencing. In addition, we also evaluate the correlation of mutations detected from this study with progression-free survival (PFS).
Materials and methods
A total of 56 HR-MBC patients were enrolled. Cell-free DNA (cfDNA) was extracted from plasma and sequenced by using Oncomine Breast cancer cfDNA assay in this study.
Result
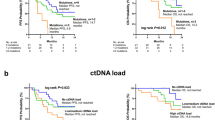
Concentration of cfDNA is correlated with a number of metastatic organs and serum CEA levels (Spearman’s rank correlation p = 0.0018, p = 0.0015 respectively). Cases with high cfDNA levels (≥ 2.6 ng/μl of plasma) showed worse progression-free survival (PFS) and overall survival compared with cases with low cfDNA levels (p = 0.043 and 0.046, respectively). Among these patients, 29 patients (51.7%) have TP53 mutations, 12 patients (30.3%) have PIK3CA mutations, and 9 patients (16.0%) have ESR1 mutations. Acquisition of ESR1 mutation increased according to the lines of hormone therapy. In addition, patients with ESR1 mutation showed shorter PFS than those without mutation (log-rank p = 0.047). In the multivariate analysis, ESR1 mutation and cfDNA concentration were significant for PFS (p = 0.027 and 0.006, respectively). In conclusion, assessment of ESR1 mutation and cfDNA concentration could be useful in predicting prognosis for HR-MBCs.





Similar content being viewed by others
References
O'Shaughnessy J (2005) Extending survival with chemotherapy in metastatic breast cancer. Oncologist 10(Suppl 3):20–29. https://doi.org/10.1634/theoncologist.10-90003-20
Toss A, Cristofanilli M (2015) Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res 17:60. https://doi.org/10.1186/s13058-015-0560-9
Mouridsen H, Gershanovich M, Sun Y, Perez-Carrion R, Boni C, Monnier A, Apffelstaedt J, Smith R, Sleeboom HP, Jaenicke F, Pluzanska A, Dank M, Becquart D, Bapsy PP, Salminen E, Snyder R, Chaudri-Ross H, Lang R, Wyld P, Bhatnagar A (2003) Phase III study of letrozole versus tamoxifen as first-line therapy of advanced breast cancer in postmenopausal women: analysis of survival and update of efficacy from the International Letrozole Breast Cancer Group. J Clin Oncol 21(11):2101–2109. https://doi.org/10.1200/JCO.2003.04.194
Johnston SR, Kilburn LS, Ellis P, Dodwell D, Cameron D, Hayward L, Im YH, Braybrooke JP, Brunt AM, Cheung KL, Jyothirmayi R, Robinson A, Wardley AM, Wheatley D, Howell A, Coombes G, Sergenson N, Sin HJ, Folkerd E, Dowsett M, Bliss JM, So FEAi (2013) Fulvestrant plus anastrozole or placebo versus exemestane alone after progression on non-steroidal aromatase inhibitors in postmenopausal patients with hormone-receptor-positive locally advanced or metastatic breast cancer (SoFEA): a composite, multicentre, phase 3 randomised trial. Lancet Oncol 14(10):989–998. https://doi.org/10.1016/S1470-2045(13)70322-X
Hortobagyi GN (1998) Treatment of breast cancer. N Engl J Med 339(14):974–984. https://doi.org/10.1056/NEJM199810013391407
Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, Hudis C, Chen D, Taran T, Hortobagyi G, Greene G, Berger M, Baselga J, Chandarlapaty S (2013) ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45(12):1439–1445. https://doi.org/10.1038/ng.2822
Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D, Rizel S, Klein B, Rubinek T, Wolf I (2013) D538G mutation in estrogen receptor-alpha: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res 73(23):6856–6864. https://doi.org/10.1158/0008-5472.CAN-13-1197
Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, Ding L, Griffith OL, Miller C, Larson D, Fulton RS, Harrison M, Mooney T, McMichael JF, Luo J, Tao Y, Goncalves R, Schlosberg C, Hiken JF, Saied L, Sanchez C, Giuntoli T, Bumb C, Cooper C, Kitchens RT, Lin A, Phommaly C, Davies SR, Zhang J, Kavuri MS, McEachern D, Dong YY, Ma C, Pluard T, Naughton M, Bose R, Suresh R, McDowell R, Michel L, Aft R, Gillanders W, DeSchryver K, Wilson RK, Wang S, Mills GB, Gonzalez-Angulo A, Edwards JR, Maher C, Perou CM, Mardis ER, Ellis MJ (2013) Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 4(6):1116–1130. https://doi.org/10.1016/j.celrep.2013.08.022
Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, Hayes DF, Chugh R, Kunju LP, Talpaz M, Schott AF, Chinnaiyan AM (2013) Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45(12):1446–1451. https://doi.org/10.1038/ng.2823
Tamrazi A, Carlson KE, Rodriguez AL, Katzenellenbogen JA (2005) Coactivator proteins as determinants of estrogen receptor structure and function: spectroscopic evidence for a novel coactivator-stabilized receptor conformation. Mol Endocrinol 19(6):1516–1528. https://doi.org/10.1210/me.2004-0458
Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, De Stanchina E, Carlson KE, Martin TS, Uddin S, Li Z, Fanning S, Katzenellenbogen JA, Greene G, Baselga J, Chandarlapaty S (2017) Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov 7(3):277–287. https://doi.org/10.1158/2159-8290.CD-15-1523
Rouhanifard SH, Mellis IA, Dunagin M, Bayatpour S, Jiang CL, Dardani I, Symmons O, Emert B, Torre E, Cote A, Sullivan A, Stamatoyannopoulos JA, Raj A (2019) Amendments: author correction: ClampFISH detects individual nucleic acid molecules using click chemistry-based amplification. Nat Biotechnol 37(1):102. https://doi.org/10.1038/nbt0119-102b
Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A, Dive C (2010) Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 120(2):461–467. https://doi.org/10.1007/s10549-010-0747-9
Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC (2015) EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 90(3):509–515. https://doi.org/10.1016/j.lungcan.2015.10.004
Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, Massard C, Levy C, Arnedos M, Lacroix-Triki M, Garrabey J, Boursin Y, Deloger M, Fu Y, Commo F, Scott V, Lacroix L, Dieci MV, Kamal M, Dieras V, Goncalves A, Ferrerro JM, Romieu G, Vanlemmens L, Mouret Reynier MA, Thery JC, Le Du F, Guiu S, Dalenc F, Clapisson G, Bonnefoi H, Jimenez M, Le Tourneau C, Andre F (2016) Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med 13(12):e1002201. https://doi.org/10.1371/journal.pmed.1002201
Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, Hadfield J, May AP, Caldas C, Brenton JD, Rosenfeld N (2012) Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 4 (136):136ra168. doi:10.1126/scitranslmed.3003726
Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B (2003) Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA 100(15):8817–8822. https://doi.org/10.1073/pnas.1133470100
Vogelstein B, Kinzler KW (1999) Digital PCR. Proc Natl Acad Sci USA 96(16):9236–9241. https://doi.org/10.1073/pnas.96.16.9236
Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K, Blons H, Bouche O, Landi B, Hutchison JB, Laurent-Puig P (2013) Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 59(12):1722–1731. https://doi.org/10.1373/clinchem.2013.206359
Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17(4):223–238. https://doi.org/10.1038/nrc.2017.7
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11(5):759–769. https://doi.org/10.1111/j.1755-0998.2011.03024.x
Masunaga N, Kagara N, Motooka D, Nakamura S, Miyake T, Tanei T, Naoi Y, Shimoda M, Shimazu K, Kim SJ, Noguchi S (2018) Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Treat 167(1):49–58. https://doi.org/10.1007/s10549-017-4487-y
Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda S, Perez-Lopez R, Dolling D, Robinson DR, Sandhu S, Fowler G, Ebbs B, Flohr P, Seed G, Rodrigues DN, Boysen G, Bertan C, Atkin M, Clarke M, Crespo M, Figueiredo I, Riisnaes R, Sumanasuriya S, Rescigno P, Zafeiriou Z, Sharp A, Tunariu N, Bianchini D, Gillman A, Lord CJ, Hall E, Chinnaiyan AM, Carreira S, de Bono JS (2017) Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 7(9):1006–1017. https://doi.org/10.1158/2159-8290.CD-17-0261
Miao Y, Fan Y, Zhang L, Ma T, Li R (2019) Clinical value of plasma cfDNA concentration and integrity in breast cancer patients. Cell Mol Biol (Noisy-le-grand) 65 (6):64–72
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA, Jr. (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224):224. https://doi.org/10.1126/scitranslmed.3007094
Pharoah PD, Day NE, Caldas C (1999) Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 80(12):1968–1973. https://doi.org/10.1038/sj.bjc.6690628
Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12(4):1157–1167. https://doi.org/10.1158/1078-0432.CCR-05-1029
Williams AB, Schumacher B (2016) p53 in the DNA-damage-repair process. Cold Spring Harb Perspect Med 6(5):a026070. https://doi.org/10.1101/cshperspect.a026070
Kim JY, Lee E, Park K, Park WY, Jung HH, Ahn JS, Im YH, Park YH (2017) Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes. Oncotarget 8(17):27997–28007. https://doi.org/10.18632/oncotarget.15881
Meric-Bernstam F, Zheng X, Shariati M, Damodaran S, Wathoo C, Brusco L, Demirhan ME, Tapia C, Eterovic AK, Basho RK, Ueno NT, Janku F, Sahin A, Rodon J, Broaddus R, Kim TB, Mendelsohn J, Mills Shaw KR, Tripathy D, Mills GB, Chen K (2018) Survival outcomes by TP53 mutation status in metastatic breast cancer. JCO Precis Oncol 2(2018):1–15. https://doi.org/10.1200/PO.17.00245
Miron A, Varadi M, Carrasco D, Li H, Luongo L, Kim HJ, Park SY, Cho EY, Lewis G, Kehoe S, Iglehart JD, Dillon D, Allred DC, Macconaill L, Gelman R, Polyak K (2010) PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res 70(14):5674–5678. https://doi.org/10.1158/0008-5472.CAN-08-2660
Zardavas D, Te Marvelde L, Milne RL, Fumagalli D, Fountzilas G, Kotoula V, Razis E, Papaxoinis G, Joensuu H, Moynahan ME, Hennessy BT, Bieche I, Saal LH, Stal O, Iacopetta B, Jensen JD, O'Toole S, Lopez-Knowles E, Barbaraeschi M, Noguchi S, Azim HA Jr, Lerma E, Bachelot T, Wang Q, Perez-Tenorio G, Can de Velde CJH, Rea DW, Sabine V, Bartlett JMS, Sotiriou C, Michiels S, Loi S (2018) Tumor PIK3CA Genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol 36(10):981–990. https://doi.org/10.1200/JCO.2017.74.8301
Allouchery V, Beaussire L, Perdrix A, Sefrioui D, Augusto L, Guillemet C, Sarafan-Vasseur N, Di Fiore F, Clatot F (2018) Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Cancer Res 20(1):40. https://doi.org/10.1186/s13058-018-0968-0
Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S, Jiang J, Bartlett CH, Koehler M, Dowsett M, Bliss JM, Johnston SR, Turner NC (2016) Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol 34(25):2961–2968. https://doi.org/10.1200/JCO.2016.67.3061
Acknowledgements
We thank Aya Imai and Fumie Sakamoto for their technical support. We would like to convey our gratitude to Dr. Toru Hirota for his constructive suggestions in this study. This study is supported by internal funding of Japanese Foundation for Cancer Research and Novartis Research Grant.
Funding
This study was funded by the Internal funds of JFCR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Shunji Takahashi has received research grant for other studies from AstraZeneca, Chugai Pharmaceutical, Eisai, Novartis, Bayer, Taiho, Daiichi-sankyo, and MSD. Yoshinori Ito has received research Grant for other studies from AstraZeneca, Chugai Pharmaceutical, Novartis, EPS, Daiichi-sankyo, Lilly, Kyowa Hakko Kirin, Covance, A2 healthcare, IQVIA, and MSD. Shinji Ono has received research Grant for other studies from Chugai, Eisai, Novartis, Lilly, Pfizer, Taiho, AstraZeneca, and Kyowa Hakko Kirin. Takayuki Ueno received honoraria from Chugai Pharmaceutical, Eisai, Novartis, and Astra Zeneca. Yusuke Nakamura is in an advisory role of Onco Therapy Science, Inc. The other authors declare that they do not have a financial relationship with the organizations that sponsored the research.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shibayama, T., Low, SK., Ono, M. et al. Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res Treat 180, 331–341 (2020). https://doi.org/10.1007/s10549-019-05512-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05512-5