Abstract
Purpose
PALB2 variants have been scarcely described in Argentinian and Latin-American reports. In this study, we describe molecular and clinical characteristics of PALB2 mutations found in multi-gene panels (MP) from breast-ovarian cancer (BOC) families in different institutions from Argentina.
Methods
We retrospectively identified PALB2 pathogenic (PV) and likely pathogenic (LPV) variants from a cohort of 1905 MP results, provided by one local lab (Heritas) and SITHER (Hereditary Tumor Information System) public database. All patients met hereditary BOC clinical criteria for testing, according to current guidelines.
Results
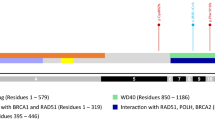
The frequency of PALB2 mutations is 2.78% (53/1905). Forty-eight (90.5%) are PV and five (9.5%) are LPV. Most of the 18 different mutations (89%) are nonsense and frameshift types and 2 variants are novel. One high-rate recurrent PV (Y551*) is present in 43% (23/53) of the unrelated index cases. From the 53 affected carriers, 94% have BC diagnosis with 14% of bilateral cases. BC phenotype is mainly invasive ductal (78%) with 62% of hormone-receptor positive and 22% of triple negative tumors. Self-reported ethnic background of the cohort is West European (66%) and native Latin-American (20%) which is representative of Buenos Aires and other big urban areas of the country.
Conclusion
This is the first report describing molecular and clinical characteristics of PALB2 carriers in Argentina. Frequency of PALB2 PV in Argentinian HBOC families is higher than in other reported populations. Y551* is a recurrent mutation that seems to be responsible for almost 50% of PALB2 cases.

Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in SITHER public database (http://www.inc.gob.ar/sither) and in this article with its supplementary information files.
Notes
European descendants born in the Americas [45]
References
Antoniou AC, Casadei S, Heikkinen T et al (2014) Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371(6):497–506. https://doi.org/10.1056/NEJMoa1400382
Yang X, Leslie G, Doroszuk A et al (2020) Cancer risks associated with Germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol 38(7):674–685. https://doi.org/10.1200/JCO.19.01907
Breast Cancer Association Consortium, Dorling L, Carvalho S, Allen J et al (2021) Breast cancer risk genes—association analysis in more than 113,000 women. N Engl J Med 384(5):428–439. https://doi.org/10.1056/NEJMoa1913948
Hu C, Hart SN, Gnanaolivu R et al (2021) A population-based study of genes previously implicated in breast cancer. N Engl J Med 384(5):440–451. https://doi.org/10.1056/NEJMoa2005936
Kurian AW, Hughes E, Handorf EA, Gutin A, Allen B, Hartman AR, Hall MJ (2017) Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol 1:1–12. https://doi.org/10.1200/PO.16.00066
Tischkowitz M, Balmaña J, Foulkes WD, James P, Ngeow J, Schmutzler R, Voian N, Wick MJ, Stewart DR, Pal T, Professional Practice ACMG, Committee G (2021) Management of individuals with germline variants in PALB2: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med 23(8):1416–1423. https://doi.org/10.1038/s41436-021-01151-8
Hauke J, Horvath J, Groß E et al (2018) Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med 7(4):1349–1358. https://doi.org/10.1002/cam4.1376
Kluska A, Balabas A, Piatkowska M, Czarny K, Paczkowska K, Nowakowska D, Mikula M, Ostrowski J (2017) PALB2 mutations in BRCA1/2-mutation negative breast and ovarian cancer patients from Poland. BMC Med Genomics 10(1):14. https://doi.org/10.1186/s12920-017-0251-8
Nguyen-Dumont T, Hammet F, Mahmoodi M, Tsimiklis H, Teo ZL, Li R, Pope BJ, Terry MB, Buys SS, Daly M, Hopper JL, Winship I, Goldgar DE, Park DJ, Southey MC (2015) Mutation screening of PALB2 in clinically ascertained families from the Breast Cancer Family Registry. Breast Cancer Res Treat 149(2):547–554. https://doi.org/10.1007/s10549-014-3260-8
Erkko H, Xia B, Nikkilä J et al (2007) A recurrent mutation in PALB2 in Finnish cancer families. Nature 446(7133):316–319. https://doi.org/10.1038/nature05609
Wojcik P, Jasiowka M, Strycharz E, Sobol M, Hodorowicz-Zaniewska D, Skotnicki P, Byrski T, Blecharz P, Marczyk E, Cedrych I, Jakubowicz J, Lubiński J, Sopik V, Narod S, Pierzchalski P (2016) Recurrent mutations of BRCA1, BRCA2 and PALB2 in the population of breast and ovarian cancer patients in Southern Poland. Hered Cancer Clin Pract 14:5. https://doi.org/10.1186/s13053-016-0046-5
Zhou J, Wang H, Fu F, Li Z, Feng Q, Wu W, Liu Y, Wang C, Chen Y (2020) Spectrum of PALB2 germline mutations and characteristics of PALB2-related breast cancer: screening of 16,501 unselected patients with breast cancer and 5890 controls by next-generation sequencing. Cancer 126(14):3202–3208. https://doi.org/10.1002/cncr.32905
Cerretini R, Mercado G, Morganstein J, Schiaffi J, Reynoso M, Montoya D, Valdéz R, Narod SA, Akbari MR (2019) Germline pathogenic variants in BRCA1, BRCA2, PALB2 and RAD51C in breast cancer women from Argentina. Breast Cancer Res Treat 178(3):629–636. https://doi.org/10.1007/s10549-019-05411-9
SITHER -Sistema de Información de Tumores Hereditarios- website Instituto Nacional del Cáncer de Argentina. http://www.inc.gob.ar/sither. Accessed 11 March 2022
Daly MB, Pal T, Berry MP et al (2021) Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Comp Cancer Netw JNCCN 19(1):77–102. https://doi.org/10.6004/jnccn.2021.0001
Nuñez L (2019) Protocolo de paneles multigenéticos en cáncer hereditario. Instituto Nacional del Cáncer de Argentina. https://bancos.salud.gob.ar/recurso/protocolo-de-paneles-multigeneticos-en-cancer-hereditario. Accessed 11 Mar 2022
Huo D, Melkonian S, Rathouz PJ, Khramtsov A, Olopade OI (2011) Concordance in histological and biological parameters between first and second primary breast cancers. Cancer 117(5):907–915. https://doi.org/10.1002/cncr.25587
Bernstein JL, Lapinski RH, Thakore SS, Doucette JT, Thompson WD (2003) The descriptive epidemiology of second primary breast cancer. Epidemiology (Cambridge, MA) 14(5):552–558. https://doi.org/10.1097/01.ede.0000072105.39021.6d
Miller DT, Lee K, Chung WK et al (2021) ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Supp table 1. Genet Med 23:1381–1390. https://doi.org/10.1038/s41436-021-01172-3
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Resource C (2021) Sequence Variant Interpretation – ClinGen | Clinical Genome Resource. https://clinicalgenome.org/working-groups/sequence-variant-interpretation/. Accessed 16 Dec 2021
Garrett A, Callaway A, Durkie M et al (2020) Cancer Variant Interpretation Group UK (CanVIG-UK): an exemplar national subspecialty multidisciplinary network. J Med Genet 57(12):829–834. https://doi.org/10.1136/jmedgenet-2019-106759
Chunn LM, Nefcy DC, Scouten RW, Tarpey RP, Chauhan G, Lim MS, Elenitoba-Johnson K, Schwartz SA, Kiel MJ (2020) Mastermind: a comprehensive genomic association search engine for empirical evidence curation and genetic variant interpretation. Front Genet 11:577152. https://doi.org/10.3389/fgene.2020.577152
Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, Hallberg E, Moore R, Thomas A, Lilyquist J, Feng B, McFarland R, Pesaran T, Huether R, LaDuca H, Chao EC, Goldgar DE, Dolinsky JS (2017) Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol 3(9):1190–1196. https://doi.org/10.1001/jamaoncol.2017.0424
Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, Sharma L, Saam J, Lancaster J, Daly MB (2017) A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123(10):1721–1730. https://doi.org/10.1002/cncr.30498
Thompson ER, Gorringe KL, Rowley SM, Wong-Brown MW, McInerny S, Li N, Trainer AH, Devereux L, Doyle MA, Li J, Lupat R, Delatycki MB, Investigators LifePool, Mitchell G, James PA, Scott RJ, Campbell IG (2015) Prevalence of PALB2 mutations in Australian familial breast cancer cases and controls. Breast Cancer Res BCR 17(1):111. https://doi.org/10.1186/s13058-015-0627-7
Preobrazhenskaya EV, Shleykina AU, Gorustovich OA, Martianov AS, Bizin IV, Anisimova EI, Sokolova TN, Chuinyshena SA, Kuligina ES, Togo AV, Belyaev AM, Ivantsov AO, Sokolenko AP, Imyanitov EN (2021) Frequency and molecular characteristics of PALB2-associated cancers in Russian patients. Int J Cancer 148(1):203–210. https://doi.org/10.1002/ijc.33317
Vietri MT, Caliendo G, Schiano C, Casamassimi A, Molinari AM, Napoli C, Cioffi M (2015) Analysis of PALB2 in a cohort of Italian breast cancer patients: identification of a novel PALB2 truncating mutation. Fam Cancer 14(3):341–348. https://doi.org/10.1007/s10689-015-9786-z
Singh J, Thota N, Singh S et al (2018) Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations. Breast Cancer Res Treat 170(1):189–196. https://doi.org/10.1007/s10549-018-4726-x
Lerner-Ellis J, Donenberg T, Ahmed H, George S, Wharfe G, Chin S, Lowe D, Royer R, Zhang S, Narod S, Hurley J, Akbari MR (2017) A high frequency of PALB2 mutations in Jamaican patients with breast cancer. Breast Cancer Res Treat 162(3):591–596. https://doi.org/10.1007/s10549-017-4148-1
Weitzel JN, Neuhausen SL, Adamson A, Tao S, Ricker C, Maoz A, Rosenblatt M, Nehoray B, Sand S, Steele L, Unzeitig G, Feldman N, Blanco AM, Hu D, Huntsman S, Castillo D, Haiman C, Slavin T, Ziv E (2019) Pathogenic and likely pathogenic variants in PALB2, CHEK2, and other known breast cancer susceptibility genes among 1054 BRCA-negative Hispanics with breast cancer. Cancer 125(16):2829–2836. https://doi.org/10.1002/cncr.32083
Cock-Rada AM, Ossa CA, Garcia HI, Gomez LR (2018) A multi-gene panel study in hereditary breast and ovarian cancer in Colombia. Fam Cancer 17(1):23–30. https://doi.org/10.1007/s10689-017-0004-z
Ren M, Orozco A, Shao K, Albanez A, Ortiz J, Cao B, Wang L, Barreda L, Alvarez CS, Garland L, Wu D, Chung CC, Wang J, Frone M, Ralon S, Argueta V, Orozco R, Gharzouzi E, Dean M (2021) Germline variants in hereditary breast cancer genes are associated with early age at diagnosis and family history in Guatemalan breast cancer. Breast Cancer Res Treat 189(2):533–539. https://doi.org/10.1007/s10549-021-06305-5
de Souza Timoteo AR, Gonçalves A, Sales L, Albuquerque BM, de Souza J, de Moura P, de Aquino M, Agnez-Lima LF, Lajus T (2018) A portrait of germline mutation in Brazilian at-risk for hereditary breast cancer. Breast Cancer Res Treat 172(3):637–646. https://doi.org/10.1007/s10549-018-4938-0
Sandoval RL, Leite A, Barbalho DM, Assad DX, Barroso R, Polidorio N, Dos Anjos CH, de Miranda AD, Ferreira A, Fernandes G, Achatz MI (2021) Germline molecular data in hereditary breast cancer in Brazil: lessons from a large single-center analysis. PLoS ONE 16(2):e0247363. https://doi.org/10.1371/journal.pone.0247363
da Costa E, Silva Carvalho S, Cury NM, Brotto DB, de Araujo LF, Rosa R, Texeira LA, Plaça JR, Marques AA, Peronni KC, Ruy PC, Molfetta GA, Moriguti JC, Carraro DM, Palmero EI, Ashton-Prolla P, de Faria Ferraz VE, Silva WA Jr (2020) Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: analysis of a 21 gene panel in the Brazilian population. BMC Med Genomics 13(1):21. https://doi.org/10.1186/s12920-019-0652-y
Adaniel C, Salinas F, Donaire JM, Bravo ME, Peralta O, Paredes H, Aliaga N, Sola A, Neira P, Behnke C, Rodriguez T, Torres S, Lopez F, Hurtado C (2019) Non-BRCA1/2 variants detected in a high-risk Chilean cohort with a history of breast and/or ovarian cancer. J Glob Oncol 5:1–14. https://doi.org/10.1200/JGO.18.00163
Leyton Y, Gonzalez-Hormazabal P, Blanco R, Bravo T, Fernandez-Ramires R, Morales S, Landeros N, Reyes JM, Peralta O, Tapia JC, Gomez F, Waugh E, Ibañez G, Pakomio J, Grau G, Jara L (2015) Association of PALB2 sequence variants with the risk of familial and early-onset breast cancer in a South-American population. BMC Cancer 15:30. https://doi.org/10.1186/s12885-015-1033-3
Solano AR, Mele PG, Jalil FS, Liria NC, Podesta EJ, Gutiérrez LG (2021) Study of the genetic variants in BRCA1/2 and Non-BRCA genes in a population-based cohort of 2155 breast/ovary cancer patients, including 443 triple-negative breast cancer patients, in Argentina. Cancers 13(11):2711. https://doi.org/10.3390/cancers13112711
Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39(2):159–161. https://doi.org/10.1038/ng1942
Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC (2011) Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Can Res 71(6):2222–2229. https://doi.org/10.1158/0008-5472.CAN-10-3958
Blanco A, de la Hoya M, Osorio A et al (2013) Analysis of PALB2 gene in BRCA1/BRCA2 negative Spanish hereditary breast/ovarian cancer families with pancreatic cancer cases. PLoS ONE 8(7):e67538. https://doi.org/10.1371/journal.pone.0067538
Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, Lilyquist J, Na J, Moore R, Antwi SO, Bamlet WR, Chaffee KG, DiCarlo J, Wu Z, Samara R, Kasi PM, McWilliams RR, Petersen GM, Couch FJ (2018) Association between inherited Germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 319(23):2401–2409. https://doi.org/10.1001/jama.2018.6228
National Center for Biotechnology Information. ClinVar; [VCV000001243.19], https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000001243.19. Accessed 8 Nov 2021
Roberts AP (2006) The odyssey of criollo. In: Lang PL (ed) Studies in contact linguistics. Lang, New York, pp 6–7
Ramírez-Calvo M, García-Casado Z, Fernández-Serra A, de Juan I, Palanca S, Oltra S, Soto JL, Castillejo A, Barbera VM, Juan-Fita MJ, Segura Á, Chirivella I, Sánchez AB, Tena I, Chaparro C, Salas D, López-Guerrero JA (2019) Implementation of massive sequencing in the genetic diagnosis of hereditary cancer syndromes: diagnostic performance in the Hereditary Cancer Programme of the Valencia Community (FamCan-NGS). Hered Cancer Clin Pract 17:3. https://doi.org/10.1186/s13053-019-0104-x
National Center for Biotechnology Information. ClinVar; [VCV000481035.8], https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000481035.8. Accessed 10 Nov 2021
National Center for Biotechnology Information. ClinVar; [VCV000141560.11], https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000141560.11. Accessed 10 Nov 2021
Mucaki EJ, Caminsky NG, Perri AM, Lu R, Laederach A, Halvorsen M, Knoll JH, Rogan PK (2016) A unified analytic framework for prioritization of non-coding variants of uncertain significance in heritable breast and ovarian cancer. BMC Med Genomics 9:19. https://doi.org/10.1186/s12920-016-0178-5
Ng PS, Boonen RA, Wijaya E et al (2021) Characterisation of protein-truncating and missense variants in PALB2 in 15 768 women from Malaysia and Singapore. J Med Genet. https://doi.org/10.1136/jmedgenet-2020-107471
Corach D, Lao O, Bobillo C, van Der Gaag K, Zuniga S, Vermeulen M, van Duijn K, Goedbloed M, Vallone PM, Parson W, de Knijff P, Kayser M (2010) Inferring continental ancestry of Argentineans from Autosomal, Y-chromosomal and mitochondrial DNA. Ann Hum Genet 74(1):65–76. https://doi.org/10.1111/j.1469-1809.2009.00556.x
Wang S, Ray N, Rojas W et al (2008) Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet 4(3):e1000037. https://doi.org/10.1371/journal.pgen.1000037
Luisi P, García A, Berros JM et al (2020) Fine-scale genomic analyses of admixed individuals reveal unrecognized genetic ancestry components in Argentina. PLoS ONE 15(7):e0233808. https://doi.org/10.1371/journal.pone.0233808
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
This research study was conducted retrospectively from data obtained for clinical purposes and public databases. Informed consent was obtained from patients for both germline genetic testing and anonymous use of clinical information at the time of the sample collection.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gonzalez, A., Del Greco, F., Vargas-Roig, L. et al. PALB2 germline mutations in a multi-gene panel testing cohort of 1905 breast-ovarian cancer patients in Argentina. Breast Cancer Res Treat 194, 403–412 (2022). https://doi.org/10.1007/s10549-022-06620-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-022-06620-5