Abstract
Objective
The heterotrophic marine microalga, Schizochytrium mangrovei PQ6, synthesizes large amounts of polyunsaturated fatty acids (PUFAs) with possible nutritional applications. We characterized the transcriptome of S. mangrovei PQ6, focusing on lipid metabolism pathways throughout growth.
Result
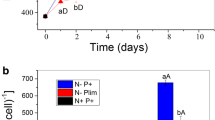
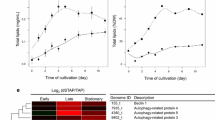
Cell growth, total lipid, and docosahexaenoic acid (DHA, 22:6n-3) contents of S. mangrovei PQ6 in 500 ml batch cultures rapidly increased on day 1 in cultivation and reached their maximum levels on day 5. Maximum lipid accumulation in 500 ml batch cultures occurred on day 5, with total lipid and DHA contents reaching 33.2 ± 1.25% of dry cell weight (DCW) and 136 mg/g DCW, respectively. 11,025 unigenes, 28,617 unigenes and 18,480 unigenes from the transcriptomes of samples collected on day 1, 3, and 5 in cultivation were identified, respectively. These unigenes of the three samples were further assembled into 30,782 unigenes with an average size of 673 bp and N50 of 950 bp, and a total of 9,980 unigenes were annotated in public protein databases. 93 unigenes involved in lipid metabolism in which expression patterns corresponded with total lipid and DHA accumulation patterns were identified.
Conclusion
The possible roles of PUFAs pathways, such as those mediated by fatty acid synthase, polyketide synthase, and desaturase/elongase, co-exist in S. mangrovei PQ6.



Similar content being viewed by others
References
Gahlan P, Singh HR, Shankar R et al (2012) De novo sequencing and characterization of Picrorhiza kurrooa transcriptome at two temperatures showed major transcriptome adjustments. BMC Genom 13:126
Gardner MJ, Hall N, Fung E et al (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511
Haas BJ, Papanicolaou A, Yassour M et al (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Hauvermale A, Kuner J, Rosenzweig B et al (2006) Fatty acid production in Schizochytrium sp.: Involvement of a polyunsaturated fatty acid synthase and a type I fatty acid synthase. Lipids 41:739–747
Hoang MH, Ha NC, Thom LT et al (2014) Extraction of squalene as value-added product from the residual biomass of Schizochytrium mangrovei PQ6 during biodiesel producing process. J Biosci Bioeng 118:632–639
Hong DD, Anh HTL, Thu NTH (2011) Study on biological characteristics of heterotrophic marine microalga—Schizochytrium mangrovei Pq6 isolated from Phu Quoc Island, Kien Giang Province, Vietnam1. J Phycol 47:944–954
Hou R, Bao Z, Wang S et al (2011) Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One 6:e21560
Lippmeier JC, Crawford KS, Owen CB et al (2009) Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids 44:621–630
Meyer A, Cirpus P, Ott C et al (2003) Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a delta4-fatty acyl group desaturase. Biochemistry (Mosc) 42:9779–9788
Mu HN, Li HG, Wang LG et al (2014) Transcriptome sequencing and analysis of sweet osmanthus (Osmanthus fragrans Lour.). Genes Genomics 36:777–788
Okamoto N, Inouye I (2005) A secondary symbiosis in progress? Science 310:287
Qiu X (2003) Biosynthesis of docosahexaenoic acid (DHA, 22:6–4,7,10,13,16,19): two distinct pathways. Prostagland Leukot Essent Fatty Acids 68:181–186
Raghukumar S (2008) Thraustochytrid marine protists: production of PUFAs and other emerging technologies. Mar Biotechnol 10:631–640
Rismani-Yazdi H, Haznedaroglu BZ, Bibby K, Peccia J (2011) Transcriptome sequencing and annotation of the microalgae Dunaliella tertiolecta: pathway description and gene discovery for production of next-generation biofuels. BMC Genom 12:148
Velmurugan N, Sathishkumar Y, Yim SS et al (2014) Study of cellular development and intracellular lipid bodies accumulation in the thraustochytrid Aurantiochytrium sp. KRS101. Bioresour Technol 161:149–154
Wang X, Zhou C, Yang X et al (2015) De novo transcriptome analysis of Warburgia ugandensis to identify genes involved in terpenoids and unsaturated fatty acids biosynthesis. PLoS One 10:e0135724
Zenoni S, Ferrarini A, Giacomelli E et al (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152:1787–1795
Zheng M, Tian J, Yang G et al (2013) Transcriptome sequencing, annotation and expression analysis of Nannochloropsis sp. at different growth phases. Gene 523:117–121
Acknowledgments
This study was supported by the National Research and Development Program of Biotechnology KC.04/11-15, the Ministry of Science and Technology, Vietnam under Grant number KC.04.20/11-15. We are grateful for the use of the facilities of National Key Laboratory, Institute of Biotechnology, VAST.
Supporting information
Methods—A-Transmission electron microscopy; B-De novo assembly of the S. mangrovei PQ6 transcriptome; C-Functional annotation; D-Differential gene expression analysis.
Supplementary Table 1—Primers used for qPCR.
Supplementary Table 2—Over representation analysis of specific enriched significant GO-slim terms on three cluster based on FPKM expression score.
Supplementary Table 3—Enzymes involved in fatty acid biosynthesis and metabolism identified by annotation of S. mangrovei PQ6 transcriptomes at different cultivation periods.
Supplementary Figure 1—The normal length distribution for BLASTX-annotated final unigene set (a) and high number of small-size unigenes for unannotated final unigene set (b).
Supplementary Figure 2—Gene Ontology (GO), Clusters of Orthologous Group (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) categories of assembled unigenes of S. mangrovei PQ6.
Supplementary Figure 3—Heat map plot of 1007 DEGs using the hierarchical clustering method from different growth stages
Author information
Authors and Affiliations
Corresponding author
Additional information
Minh Hien Hoang and Cuong Nguyen have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hoang, M.H., Nguyen, C., Pham, H.Q. et al. Transcriptome sequencing and comparative analysis of Schizochytrium mangrovei PQ6 at different cultivation times. Biotechnol Lett 38, 1781–1789 (2016). https://doi.org/10.1007/s10529-016-2165-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2165-5