Abstract
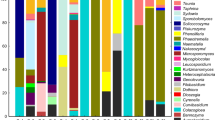
Culture-independent techniques have recently been used for evaluation of microbial diversity in the environment since it addresses the problem of unculturable microorganisms. In this study, the diversity of epiphytic yeasts from corn (Zea mays Linn.) phylloplanes in Thailand was investigated using this technique and sequence-based analysis of the D1/D2 domains of the large subunit ribosomal DNA sequences. Thirty-seven samples of corn leaf were collected randomly from 10 provinces. The DNA was extracted from leaf washing samples and the D1/D2 domains were amplified. The PCR products were cloned and then screened by colony PCR. A total of 1049 clones were obtained from 37 clone libraries. From this total, 329 clones (213 sequences) were closely related to yeast strains in the GenBank database, and they were clustered into 77 operational taxonomic units (OTUs) with a similarity threshold of 99 %. The majority of sequences (98.5 %) were classified into the phylum Basidiomycota. Sixteen known yeast species were identified. Interestingly, more than 65 % of the D1/D2 sequences obtained by this technique were suggested to be sequences from new yeast taxa. The predominant yeast sequences detected belonged to the order Ustilaginales with relative frequency of 68.0 %. The most common known yeast species detected on the leaf samples were Pseudozyma hubeiensis pro tem. and Moesziomyces antarcticus with frequency of occurrence of 24.3 and 21.6 %, respectively.








Similar content being viewed by others
References
Allen TW, Quayyum HA, Burpee LL, Buck JW (2004) Effect of foliar disease on the epiphytic yeast communities of creeping bentgrass and tall fescue. Can J Microbiol 50:853–860
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Amend A (2014) From dandruff to deep-sea vents: malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog 10:e1004277
Begerow D, Schafer AM, Kellner R, Yurkov A, Kemler M, Oberwinkler F, Bauer R (2014) Ustilaginomycotina. In: McLaughlin DJ, Spatafora JW (eds) The mycota, Vol VII. Part A: systematics and evolution, 2nd edn. Springer, Berlin, pp 295–329
Begerow D, Stoll M, Bauer R (2006) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98:906–916
Bunster L, Fokkema NJ, Schippers B (1989) Effect of surface-active Pseudomonas spp. on leaf wettability. Appl Environ Microbiol 55:1340–1345
Clement-Mathieu G, Chain F, Marchand G, Belanger RR (2008) Leaf and powdery mildew colonization by glycolipid-producing Pseudozyma species. Fungal Ecology 1:69–77
Colwell RK (2006). EstimateS: Statistical estimation of species richness and shared species from samples. Version 8. User’s Guide and application published at: http://purl.oclc.org/estimates
De Azeredo L, Gomes E, Mendonca-Hagler L, Hagler A (1998) Yeast communities associated with sugarcane in Campos, Rio de Janeiro, Brazil. International microbiology 1:205–208
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finkel OM, Burch AY, Lindow SE, Post AF, Belkin S (2011) Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl Environ Microbiol 77:7647–7655
Glushakova AM, Chernov IY (2010) Seasonal dynamics of the structure of epiphytic yeast communities. Microbiology 79:830–839
Golubev WI (2007) Mycocinogeny in smut yeast-like fungi of the genus Pseudozyma. Microbiology 76:719–722
Golubev W, Nakase T (1997) Mycocinogeny in the genus Bullera: taxonomi specificity of sensitivity to the mycocin produced by Bullera sinensis. FEMS Microbiol Lett 146:59–64
Huang R, Che HJ, Zhang J, Yang L, Jiang DH, Li GQ (2012) Evaluation of Sporidiobolus pararoseus strain YCXT3 as biocontrol agent of Botrytis cinerea on post-harvest strawberry fruits. Biol Control 62:53–63
Inácio J, Portugal L, Spencer MI, Fonseca Á (2005) Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Res 5:1167–1183
Jumpponen A, Jones KL (2010) Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol 186:496–513
Kaeberlein T, Lewis K, Epstein SS (2002) Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129
Kitamoto H, Shinozaki Y, Cao XH, Morita T, Konishi M, Tago K, Kajiwara H, Koitabashi M, Yoshida S, Watanabe T, Sameshima YY, Nakajima KT, Tsushima S (2011) Phyllosphere yeasts rapidly break down biodegradable plastics. AMB Express 1:44
Konishi M, Morita T, Fukuoka T, Imur T, Kakugawa K, Kitamoto S (2008) Efficient production of mannosylerythritol lipids with high hydrophilicity by Pseudozyma hubeiensis KM-59. Appl Microbiol Biotechnol 78:37–46
Kurtzman C, Robnett C (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Limtong S, Kaewwichian R (2015) The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann Microbiol 65:667–675
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883
Liu XZ, Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T (2015a) Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud Mycol 81:1–26
Liu XZ, Wang QM, Göker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, Bai FY (2015b) Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol 81:85–147
Molnar O, Wuczkowski M, Prillinger H (2008) Yeast biodiversity in the guts of several pests on maize; comparison of three methods: classical isolation, cloning and DGGE. Mycol Prog 7:111–123
Morita T, Koike H, Hagiwara H, Ito E, Machida M, Sato S, Habe H, Kitamoto D (2014) Genome and transcriptome analysis of the basidiomycetous yeast Pseudozyma antarctica producing extracellular glycolipids, mannosylerythritol lipids. PLoS ONE 9:e86490
Morita T, Konishi M, Fukuoka T, Imura T, Kitamoto HK, Kitamoto D (2007) Characterization of the genus Pseudozyma by the formation of glycolipid biosurfactants, mannosylerythritol lipids. FEMS Yeast Res 7:286–292
Nakase T, Takashima M, Itoh M, Fungsin B, Potacharoen W, Atthasampunna P, Komagata K (2001) Ballistoconidium-forming yeats found in the phyllosphere of Thailand. Microbiol Culture Collect 17:23–35
Nasanit R, Tangwong-o-thai A, Tantirungkij M, Limtong S (2015a) The assessment of epiphytic yeast diversity in sugarcane phyllosphere in Thailand by culture-independent method. Fungal Biol 119:1145–1157
Nasanit R, Krataithong K, Tantirungkij M, Limtong S (2015b) Assessment of epiphytic yeast diversity in rice (Oryza sativa) phyllosphere in Thailand by a culture-independent approach. Antonie Van Leeuwenhoek 107:1475–1490
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schreiber L, Krimm U, Knoll D, Sayed M, Auling G, Kroppenstedt RM (2005) Plant–microbe interactions: identification of epiphytic bacteria and their ability to alter leaf surface permeability. New Phytol 166:589–594
Seo HS, Um HJ, Min J, Rhee SK, Cho TJ, Kim YH, Lee J (2007) Pseudozyma jejuensis sp. nov., a novel cutinolytic ustilaginomycetous yeast species that is able to degrade plastic waste. FEMS Yeast Res 7:1035–1045
Slavikova E, Vadkertiova R, Vranova D (2009) Yeasts colonizing the leaves of fruit trees. Ann Microbiol 59:419–424
Takaku H, Kodaira S, Kimoto A, Nashimoto M, Takagi M (2006) Microbial communities in the garbage composting with rice hull as an amendment revealed by culture-dependent and -independent approaches. J Biosci Bioeng 101:42–50
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Voriskova J, Brabcova V, Cajthaml T, Baldrian P (2014) Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol 201:269–278
Wang QM, Jia JH, Bai FY (2006) Pseudozyma hubeiensis sp. nov. and Pseudozyma shanxiensis sp. nov., novel ustilaginomycetous anamorphic yeast species from plant leaves. Int J Syst Evol Microbiol 56:289–293
Wang QM, Groenewald M, Takashima M, Theelen B, Han PJ, Liu XZ, Boekhout T, Bai FY (2015a) Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Stud Mycol 81:27–53
Wang QM, Yurkov AM, Göker M, Lumbsch HT, Leavitt SD, Groenewald M, Theelen B, Liu XZ, Boekhout T, Bai FY (2015b) Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud Mycol 81:149–189
Wang QM, Begerow D, Groenewald M, Liu XZ, Theelen B, Bai FY, Boekhout T (2015c) Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud Mycol 81:55–83
Yoshida S, Morita T, Shinozaki Y, Watanabe T, Sameshima YY, Koitabashi M, Kitamoto D, Kitamoto H (2014) Mannosylerythritol lipids secreted by phyllosphere yeast Pseudozyma antarctica is associated with its filamentous growth and propagation on plant surfaces. Appl Microbiol Biotechnol 98:6419–6429
Yurkov A, Inacio J, Chernov I, Fonseca A (2015) Yeast biogeography and the effects of species recognition approaches: the case study of widespread basidiomycetous species from birch forests in Russia. Curr Microbiol 70:587–601
Zhang S, Schisler DA, Boehm MJ, Slininger PJ (2007) Utilization of chemical inducers of resistance and Cryptococcus flavescens OH 182.9 to reduce Fusarium head blight undergreenhouse conditions. Biol Control 42:308–315
Zimmerman NB, Vitousek PM (2012) Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc National Acad Sci 109:13022–13027
Acknowledgments
Authors are grateful to Napakhwan Imklin for PCA analysis. We also thank Kevin Maskell for English language edition. This work was supported by the Thailand Research Fund and the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nasanit, R., Jaibangyang, S., Tantirungkij, M. et al. Yeast diversity and novel yeast D1/D2 sequences from corn phylloplane obtained by a culture-independent approach. Antonie van Leeuwenhoek 109, 1615–1634 (2016). https://doi.org/10.1007/s10482-016-0762-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0762-x