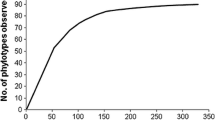
Abstract
The yeast biodiversity in the guts of several pests (Diabrotica virgifera, Helicoverpa armigera, Ostrinia nubilalis) on maize from two isolation sources was assessed by cultivation-dependent and cultivation-independent methods. These yeasts are considered to bear a potentially high biotechnological relevance due to their potential ability to degrade several mycotoxins incorporated by their hosts. The 97 isolated yeast strains showed 21 different partial sequence types of the 26S rRNA gene which could be assigned to 10 different genera. The determined genera and species are discussed in terms of the meaning of their taxonomic status or their occurrence in nature. Two cultivation-independent methods, cloning and DGGE, were compared. We propose the combination of these methods as well as the combination of both cultivation-independent and cultivation-dependent approaches, for gaining better insights into fungal biodiversity.



Similar content being viewed by others
Abbreviations
- DGGE:
-
denaturing gradient gel electrophoresis
- TGGE:
-
temperature gradient gel electrophoresis
- SSCP:
-
single strand conformation polymorphism
References
Avis TJ, Bélanger RR (2002) Mechanisms and means of detection of biocontrol activity of Pseudozyma yeasts against plant-pathogenic fungi. FEMS Yeast Res 2:5–8
Bai F-Y, Liang H-Y, Jia J-H (2000) Taxonomic relationships among the taxa in Candida guilliermondii complex, as revealed by comparative electrophoretic karyotyping. Int J Syst Evol Microbiol 50:417–422
Bai F-Y, Zhao J-H, Takashima M, Jia J-H, Boekhout T, Nakase T (2002) Reclassification of the Sporobolomyces roseus and Sporidiobolus pararoseus complexes, with the description of Sporobolomyces phaffii sp. nov. Int J Syst Evol Microbiol 52:2309–2314
Barnett JA, Payne RW, Yarrow D (2000) Yeasts: characteristics and identification, 3rd edn. Cambridge University Press, Cambridge
Begerow D, Bauer R, Boekhout T (2000) Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104:53–60
Boekhout T, Gildemacher P, Theelen B, Müller WH, Heijne B, Lutz M (2006) Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Res 6(1):63–76
Bridge P, Spooner B (2001) Soil fungi: diversity and detection. Plant Soil 232:147–154
Brysch-Herzberg M (2004) Ecology of yeasts in plant-bumblebee mutualism in Central Europe. FEMS Microbiol Ecol 50:87–100
Caetano-Anolles G, Bassam B, Gresshoff PM (1992) Primer-template interactions during DNA amplification fingerprinting with single arbitrary oligonucleotides. Mol Gen Genet 235:157–165
Cocolin L, Bisson LF, Mills DA (2000) Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett 189:81–87
Cocolin L, Aggio D, Manzano M, Cantoni C, Comi G (2002) An application of PCR-DGGE analysis to profile the yeast populations in raw milk. Int Dairy J 12:407–411
Fell JW, Statzell-Tallman A (1998) Rhodotorula F.C. Harrison. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th revised and enlarged edition. Elsevier, Amsterdam, pp 800–827
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Gadanho M, Sampaio JP (2002) Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. dairensis comb. nov. FEMS Yeast Res 2:47–58
Gardes M, Bruns TD, Taylor JW (1993) ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gomes NCM, Fagbola O, Costa R, Rumjanek NG, Buchner A, Mendona-Hagler L, Smalla K (2003) Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl Environ Microbiol 69:3758–3766
Hong SG, Lee KH, Bae KS (2002) Diversity of yeasts associated with natural environments in Korea. J Microbiol 40:55–62
Kowalchuk GA, Gerards S, Woldendorp JW (1997) Detection and characterisation of fungal infections of Ammophila arenia (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl Environ Microbiol 63:3858–3865
Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large subunit (26S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223
Kurtzman CP, Droby S (2001) Metschnikowia fructicola, a new ascosporogenic yeast with potential for biocontrol of post harvest fruit rots. Syst Appl Microbiol 24:395–399
Lachance MA, Starmer WT (1998) Ecology and yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th revised and enlarged edition. Elsevier, Amsterdam, pp 21–30
Messner R, Prillinger H, Ibl M, Himmler G (1995) Sequences of ribosomal genes and internal transcribed spacers move three plant parasitic fungi, Eremothecium ashbyi, Ashbya gossypii, and Nematospora coryli, towards to Saccharomyces cerevisiae. J Gen Appl Microbiol 41:31–42
Molnár O, Prillinger H (2005) Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst Appl Microbiol 28:717–726
Molnár O, Prillinger H (2006) Cryptococcus zeae, a new yeast species associated with Zea mays. Microbiol Res 161:347–354
Möhlenhoff P, Müller L, Gorbushina AA, Petersen K (2001) Molecular approach to the characterisation of fungal communities: methods for DNA extraction, PCR amplification and DGGE analysis of painted art objects. FEMS Microbiol Lett 195:169–173
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Myers RM, Maniatis T, Lerman S (1987) Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol 155:501–527
Novicki TJ, La Fe K, Bui L, Bui U, Geise R, Marr K, Cookson BT (2003) Genetic diversity among clinical isolates of Acremonium strictum determined during an investigation of a fatal mycosis. J Clin Microbiol 41:2623–2628
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, UK, pp 225–233
Pennanen T, Paavolainen L, Hantula J (2001) Rapid PCR-based method for the direct analysis of fungal communities in complex environmental samples. Soil Biol Biochem 33:697–699
Prakitchaiwattana CJ, Fleet GH, Heard GM (2004) Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res 4:865–877
Schabereiter-Gurtner C, Piñar G, Lubitz W, Rölleke S (2001) Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA sequence analysis. J Microbiol Methods 47:345–354
Schweigkofler W, Prillinger H (1999) Molekulare Identifizierung und phylogenetische Analyse von endophytischen und latent pathogenen Pilzen der Weinrebe. Mitt Klosterneuburg 49:65–78
Slavikova E, Vadkertiova R (2000) The occurrence of yeasts in the forest soils. J Basic Microbiol 40:207–212
Slavikova E, Vadkertiova R (2003) The diversity of yeasts in the agricultural soil. J Basic Microbiol 43:430–436
Smit E, Leeflang P, Glandorf B, Van Elsas JD, Wernars K (1999) Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol 65:2614–2621
Spencer JFT, Spencer DM (1997) Yeasts in natural and artificial habitats. Springer, Berlin Heidelberg New York
Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T (2000) Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J Clin Microbiol 38:1468–1471
Suh S-O, Blackwell M (2004) Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res 5:87–95
Suh S-O, Nguyen NH, Blackwell M (2005) Nine new Candida species near C. membranifaciens isolated from insects. Mycol Res 109:1045–1056
Takashima M, Sugita T, Shinoda T, Nakase T (2003) Three new combinations from the Cryptococcus laurentii complex: Cryptococcus aureus, Cryptococcus carnescens and Cryptococcus peneaus. Int J Syst Evol Microbiol 53:1187–1194
Vainio EJ, Hantula J (2000) Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res 104:927–936
Van Elsas JD, Duarte GF, Keijzer-Wolters A, Smit E (2000) Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J Microbiol Methods 43:133–151
Woolfolk SW, Inglis GD (2004) Microorganisms associated with field-collected Chrysoperla rufilabris (Neuroptera: Chrysopidae) adults with emphasis on the yeast symbionts. Biol Control 29:155–168
Wuczkowski M, Prillinger H (2004) Molecular identification of yeasts from soils of the alluvial forest national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Microbiol Res 159:263–275
Wuczkowski M, Sterflinger K, Kraus GF, Klug B, Prillinger H (2003) Diversity of microfungi and yeasts in soils of the alluvial zone national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Bodenkultur 54:109–117
Acknowledgements
We are grateful to Assistant Prof. Joseph Strauss and Dr. Markus Gorfer for accomplishing the cloning experiments in their laboratory. We thank to Dr. Peter Cate and Helmut Klapal who helped us to collect and identify the insects in the fields.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material
Fig. S1
Hanseniaspora uvarum, budding cells, two days on GYP agar. Bar indicates 10 μm. (GIF 36.4 kb)
Fig. S2
Tilletiopsis washingtonensis. A 2 days on cornmeal agar, note the sterigmata (arrows); B ballistoconidia. Bars indicate 10 μm. (GIF 44.9 kb)
Fig. S3
PCR-fingerprints of strains identified as Pichia guilliermondii on the basis of their rDNA sequences. Amplification primed by M13 (A), (GTG)5 (B) and V5 (C). M = length marker. CBS 2030T is the type strain of Pichia guilliermondii, CBS 5975T is the type strain of Candida xestobii and CBS 7921T is the type strain of Candida fukuyamaensis. Note the similarities of the strains identified as P. guilliermondii (HA 1658-HA 1663) and C. xestobii (HA 1664-HA 1668) amplified with all the three primers. (GIF 127 kb)
Fig. S4
PCR-fingerprints of strains identified as Aureobasidium pullulans on the basis of their rDNA sequences. Amplification primed by M13 (A), (GTG)5 (B, lanes 2–4) and V5 (B, lanes 5–7). M Length marker. CBS 584.75 is a confirmed strain of A. pullulans. (GIF 25.6 kb)
Fig. S5
PCR-fingerprints of strains identified as Candida quercitrusa. Amplification primed by M13 (lanes 2–3), (GTG)5 (lanes 4–5) and V5 (lanes 6–7). M Length marker. CBS 4412T is the type strain of C. quercitrusa. (GIF 28.4 kb)
Fig. S6
PCR-fingerprints of strains identified as Hanseniasporum uvarum. Amplification primed by M13 (lanes 1–2), (GTG)5 (lanes 3–4) and V5 (lanes 6–7). M Length marker. CBS 314T is the type strain of H. uvarum. (GIF 29.0 kb)
Fig. S7
PCR-fingerprints of strains identified as Rhodotorula glutinis. Amplification primed by M13 (lanes 1–6) and V5 (lanes 8–13). M Length marker. CBS 20T is the type strain of R. glutinis. CBS 6085T is the type strain of Rhodosporidium diobovatum. (GIF 22.0 kb)
Fig. S8
PCR-fingerprints of strains identified as Sporobolomyces coprosmae. Amplification primed by M13 (lanes 1–4) and V5 (lanes 6–9). M Length marker. CBS 7899T is the type strain of S. coprosmae. (GIF 31.9 kb)
Rights and permissions
About this article
Cite this article
Molnár, O., Wuczkowski, M. & Prillinger, H. Yeast biodiversity in the guts of several pests on maize; comparison of three methods: classical isolation, cloning and DGGE. Mycol Progress 7, 111–123 (2008). https://doi.org/10.1007/s11557-008-0558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-008-0558-0