Abstract
Biological invasions are a major threat to biodiversity and have particularly devastating impacts on island ecosystems. The New Caledonia archipelago is considered a biodiversity hotspot due to its diverse native flora. Javan rusa deer (Rusa timorensis) were introduced to New Caledonia in 1870 and the population consists of several hundred thousand individuals today. They directly threaten rare endemic species and affect the composition and structure of the vegetation. While a rusa deer management plan has identified ten priority areas for deer control operations, removing deer could be offset by the dispersal of animals back into the control areas. Here, we genotyped 628 rusa deer using 16 microsatellite markers to analyse the genetic structure of the animals in New Caledonia. We aimed to assess fine-scale genetic structure, to identify natural barriers to deer movement and to assess functional connectivity by optimising individual-based landscape resistance models. Our results suggested that rusa deer formed a single genetic population on the main New Caledonian island. The isolation-by-distance pattern suggested that female dispersal was limited, whereas males had larger dispersal distances. We assessed functional connectivity using different genetic distance metrics and all models performed poorly (mR2 ≤ 0.0043). Landscape features thus hardly affected deer movement. The characteristics of our results suggested that they were not an artefact of the colonisation history of the species. Achieving an effective reduction of deer population sizes in specific management areas will be difficult because of the deer’s high dispersal capabilities and impossible without very substantial financial investment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological invasions are a major global threat to biodiversity and can have devastating impacts on island ecosystems (Sax and Gains 2008). Alien species can thrive on islands as native species have often evolved free of strong competition, herbivory, predation or parasitism. Consequently, invaders can have a strong negative impact on their plant food, competitors or animal prey (Courchamp et al. 2003). At the same time, remote islands frequently have a high rate of endemism and make an important contribution to biodiversity relative to their size. They are thus often classified as biodiversity ‘hotspots’ (Myers et al. 2000). Eradication or management of island invasive species is thus a global conservation priority (de Wit et al. 2020).
New Caledonia, a large archipelago in the South Pacific, has been classified as a biodiversity hotspot due to its remarkably diverse native flora (Myers et al. 2000; Wulff et al. 2013). Over 3400 species of vascular plants have been recorded, around 74% of which are endemic to these islands (Morat et al. 2012; Munzinger et al. 2022). Much of this important floral diversity is severely threatened as a result of extreme environmental degradation (Wulff et al. 2013). For example, New Caledonia’s tropical dry forest contains at least 424 indigenous species of which 233 are endemic to the archipelago and 67 are restricted to this habitat (Jaffré 2009). However, this forest type is considered to be one of the most threatened tropical ecosystems worldwide (Aronson et al. 2005). Alongside intense open-cast mining, logging, wildfires and urbanisation (Pascal et al. 2008; Mansourian et al. 2018), an important threat also arises from introduced ruminant herbivores.
Javan rusa deer (Rusa timorensis) were introduced to New Caledonia in 1870 and have since become a major environmental threat. The population originated from 12 founders but underwent a dramatic expansion, reaching more than 200,000 individuals by 1940 (De Garine-Wichatitsky et al. 2003), and consisting of 250,000–370,000 individuals today (Barrière and Fort 2021). The deer are widespread in practically all terrestrial habitats, reaching particularly high densities in savannahs and tropical dry forests (De Garine-Wichatitsky et al. 2004). Through their browsing, they affect the composition and structure of the vegetation, blocking the natural regeneration process of, for instance, the tropical dry forest (Mansourian et al. 2018). Around half of their diet consists of native plants and they directly threaten some rare endemic species (De Garine-Wichatitsky et al. 2004, 2005). By overgrazing, trampling and debarking the trees, rusa deer degrade the forest undergrowth, thereby significantly contributing to soil erosion (Tramier et al. 2021).
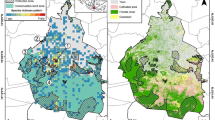
An island-wide management plan has identified ten priority areas for deer control operations with regard to the maintenance of biodiversity and ecosystem services (Fig. 1a; Tron and Barrière 2016). Since 2018, large-scale control operations, including helicopter-based hunts, have been performed in three priority areas (Anonymous 2022). However, an effective reduction in population sizes within these areas is difficult (Tron and Barrière 2016). Furthermore, even if population densities were considerably reduced to consider removal as successful, it is unclear whether natural dispersal would offset the removals. The greater the dispersal or home range of a species, the larger the spatial scale of such a source-sink effect (Woodroffe et al. 2005). While telemetry studies suggest that deer have home ranges of a few square kilometres (Moriarty 2004; Spaggiari and de Garine-Wichatitsky 2006; Amos et al. 2022), Moriarty (2004) provided evidence of dispersal of sub-adult males from a high- to a low-density deer area. Reliable information on dispersal behaviour is thus required for the implementation of effective management measures.
Geographic distribution of Rusa timorensis samples from New Caledonia. a Origin of all the samples included in this study. The yellow lines show the ten priority areas for deer control operations and a digital elevation model is used as background map. b The distribution of adult females, adult males and adults of unknown sex included in the IBD or the ResistanceGA analyses. The different geographic areas represent the different municipalities located on the main New Caledonian island (Grande Terre). Inset: Location of New Caledonia in the Indo-Pacific
The analysis of spatial genetic patterns allows inferences about animal dispersal without expansive observational or telemetry data. Genetic clustering tools allow inference of the presence of movement barriers by associating the location of abrupt genetic discontinuities with landscape elements that could disrupt dispersal (Guillot et al. 2009). Dispersal patterns can be analysed by means of an isolation-by-distance analysis. If dispersal is limited within a population, the genetic differentiation between individuals will increase with geographical distance and higher levels of philopatry will lead to a more pronounced pattern (Frantz et al. 2009, 2010). A genetic study based on limited sample size appeared to confirm a dispersal distance of 2 km or less for rusa deer on New Caledonia (de Garine-Wichatitsky et al. 2009). Finally, by statistically relating the distribution of genetic similarities among individuals to landscape characteristics, it is possible to relate gene flow patterns to landscape structure and develop rigorous empirical models of the functional connectivity of a landscape (McRae 2006; Kimmig et al. 2020).
Our overall objective was to use population and landscape genetic methods to gain a better understanding of the movement ecology of rusa deer in New Caledonia in order to optimise management strategies based on controlling deer numbers. In the light of telemetry studies, we hypothesised that (1) the presence of natural barriers as well as certain landscape features exerts a strong influence on the dispersal patterns and genetic structure of the deer and (2) the dispersal distances of the deer are inherently limited. Understanding landscape resistance and dispersal characteristics of the species will help us to understand the scale at which to operate management strategies and adjust the boundaries of the management areas to coincide with natural barriers to deer dispersal.
Materials and methods
Study area
New Caledonia has an area of 18,575 km2, making it the fourth largest archipelago in the South Pacific. The main island of Grande Terre is approximately 400 km long and 40–65 km wide (Fig. 1a; Fotsing and Dumas 2021). It is dominated along its entire length by the Chaîne Centrale mountain range with summits that reach 1628 m a.s.l. in the north and 1618 m in the south of the island (Fig. 1). The windward east coast is characterised by steep mountains rising abruptly from the sea, interrupted by narrow alluvial valleys, and the more arid west by hills and plateaus above large prairies and wetlands (Dumas 2013; Fotsing and Dumas 2021). Above 500 m a.s.l., the Chaîne Centrale is covered mostly by dense tropical rainforests. At lower altitudes on the west coast and the north of Grande Terre, land clearing and wildfires have led to savannah and scrub vegetation replacing forest habitats (Fotsing and Dumas 2021).
The south-western coastland around the capital Nouméa is highly urbanised while population densities are low elsewhere (Dumas 2013). The west coast has a shallow relief and is extensively used for agriculture, particularly cattle breeding. The majority of the forests have been converted to savannah as a result (Mansourian et al. 2018; Fotsing and Dumas 2021). Due to its narrow coastal strip and steep slopes, the eastern coast is much less developed (Fotsing and Dumas 2021). Extensive nickel reserves have resulted in intensive open-cast mining activities that currently cover 15% of New Caledonia’s total surface area (Losfeld et al. 2015; Fotsing and Dumas 2021). This type of mining has a particularly high environmental impact leading to habitat destruction and loss of local biodiversity (Pascal et al. 2008; Losfeld et al. 2015).
Sample collection
Between 2018 and 2019, we collected muscle tissue from 628 legally harvested rusa deer (Fig. 1a) and stored samples in 96% ethanol. The age category of most animals was determined based on the presence of permanent incisor teeth. In farmed animals, the first permanent incisors erupt at around 15 months (Bianchi et al. 1997). Animals without permanent incisors were thus classified as juveniles. Animals older than 15 months include both sub-adults and adults; we will refer to them as ‘adults’ for ease of reference. We also recorded the sex of most animals. The dataset consisted of 251 adult females, 196 juvenile females, 72 adult males, 41 juvenile males, 33 unsexed adults, 19 unsexed juveniles, 3 animals of unknown age class (two females, one male) and 13 animals without any information other than their geographic origin. Hunters provided the geographic origin of a sample based on official toponyms (place names), each of which is associated with geographical coordinates that correspond to the central location of a given area. New Caledonian place names are based on the history of Caledonian populations, their cultural practices and the geographical features of the island. They are sufficiently abundant to provide us with adequate geographic referencing, giving the spatial resolution of our study (Chatelier 2007).
Laboratory work and genotyping
We extracted DNA using an ammonium acetate–based salting-out procedure (Miller et al. 1988). DNA extracts were quantified using a Drop-Sense 16 spectrophotometer (Trinean, Gentbrugge, Belgium). Samples were genotyped using 16 microsatellite loci (Table 1) originally developed for red deer (Cervus elaphus), sheep (Ovis aries) and cattle (Bos taurus). Markers were selected based on the panels developed by Bonnet et al. (2002), Kuehn et al. (2003) and Pérez-Espona et al. (2008). The loci were amplified in four multiplex PCR reactions (Table 1). Each PCR contained 1 × GoTaq Master Mix (Promega, Walldorf, Germany) and 0.2 μM of each primer (except primer MM12, 0.1 μM). For multiplexes 1, 3, and 4, the PCR conditions were as follows: after a 5-min denaturation at 95 °C, the PCR consisted of 35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C (multiplexes 1 and 4) or 57 °C (multiplex 3) for 45 s and an extension at 72 °C for 45 s. The PCR was ended with a final extension for 10 min at 68 °C. In the case of the PCR for multiplex 2, the main PCR cycled consisted of a ‘touchdown’ profile, where the initial annealing temperature of 60 °C was reduced by one degree every cycle for ten cycles. This was followed by 24 cycles of annealing at 50 °C. PCRs were performed using a Mastercycler nexus cycler (Eppendorf, Hamburg, Germany) and products were separated on an ABI 3730 DNA sequencer (Applied Biosystems). All samples were genotyped at 13 loci at least.
We tested the 16 loci for heterozygote deficiency or excess (i.e. deviations from Hardy-Weinberg equilibrium (HWE)) and for linkage disequilibria among loci using a Markov chain method in genepop v3.4 (Raymond and Rousset 1995), with 10,000 dememorisation steps, 500 batches and 10,000 subsequent iterations. We tested the complete dataset (N = 628) for deviations from HWE. When analysing a dataset collected over a large geographic area, we would normally expect deviations from HWE at some loci due to the presence of distinct genetic populations in the dataset (Wahlund effect; Frankham et al. 2010). In our specific case, a number of loci in the complete dataset deviated from HWE proportions, but the dataset was not characterised by distinct genetic populations (see “Results”). Frantz et al. (2009) have shown that the presence of an isolation-by-distance gradient can cause loci to deviate from HWE, even in the absence of genetic discontinuities that are detectable by genetic clustering programs. In addition, the presence of related individuals in a dataset can lead to deviations from HWE (and to the inference of spurious genetic clusters; Bourgain et al. 2004; Anderson and Dunham 2008). To avoid deviations from HWE resulting from the presence of related individuals and isolation-by-distance (Frantz et al. 2009), we also subdivided the complete dataset arbitrarily into populations based on the municipalities of New Caledonia (Fig. 1b). We then performed the analyses only for those 13 municipalities from which ≥ 18 genetic profiles were available (range 18–127). The false discovery rate technique was used to limit false assignment of significance by chance (Verhoeven et al. 2005). We tested for the presence of null alleles using micro-checker (van Oosterhout et al. 2004).
Data analysis: Population genetics
We estimated the most-likely number of genetic clusters (K) in the complete dataset (N = 628) using structure v. 2.3.4 (Pritchard et al. 2000), conducting ten independent runs of K = 1–5 with 106 Markov chain Monte Carlo (MCMC) iterations after a burn-in of 105 iterations, using the admixture and correlated-allele-frequency models. ALPHA, the Dirichlet parameter for the degree of admixture, was allowed to vary between clusters. The most probable number of clusters was chosen based on the inferred log-likelihood values. We also analysed the complete dataset using the spatially explicit genetic clustering method implemented in the program BAPS v.6.0. (Corander et al. 2008). We performed 100 runs for K = 20. We used genetix v.4.05.2 (Belkhir et al. 2004) to estimate genetic diversity in the complete dataset in terms of number of alleles (A), as well as observed (Ho) and expected (He) heterozygosities.
We tested for the presence of an isolation-by-distance (IBD) pattern by using Spagedi v.1.5 (Hardy and Vekemans 2002) to regress pairwise estimates of Loiselle’s kinship coefficient Fij (Loiselle et al. 1995; Vekemans and Hardy 2004) against the natural logarithm of inter-individual straight-line geographic distances. The slope of this regression was tested for significant difference from zero by 10,000 permutations of the locations of the individuals. As shown by empirical data and simulations (Vekemans and Hardy 2004), Loiselle’s kinship coefficient is a relatively unbiased estimator of genetic relatedness with low sampling variance. It performs very well in detecting fine-scale genetic structure and estimating axial mother-daughter distance (see below). If samples shared the same geographic origin, we manually introduced a slight discrepancy (a few metres at most) in the coordinates to prevent them from being identical. We limited the IBD analyses to individuals defined as adults, as the inclusion of juveniles may bias results towards in a greater correlation between spatial and genetic distances (Coltman et al. 2003; Comer et al. 2005; Frantz et al. 2010). We thus performed an IBD analysis for all adult individuals (N = 356) as well as for the 251 adult females and the 72 adult males (Fig. 1b). The allele frequencies obtained from the complete data set were used in the separate analyses for males and females. A total of 10,000 randomizations of spatial locations were conducted to test the significance of the overall spatial structure (Hardy and Vekemans 2002).
Based on the IBD pattern observed with adult females (no IBD pattern with adult males; see “Results”), we used Spagedi to generate approximate estimates of the average axial mother-daughter distance (σ). When regressing relatedness coefficients on distance for diploid organisms in a two-dimensional space, σ can be inferred as σ = (− (1 − F(1)) / b4πD)0.5, where F(1) is the mean kinship coefficient between individuals in the first distance class, b the slope of the regression between pairwise spatial and genetic distances and D the effective population density (Vekemans and Hardy 2004). We used 12 spatial distance classes that Spagedi defined in such a way that the number of pairwise comparisons within each interval was constant. The first distance class had a maximum distance of 10.5 km. F(1) is usually close to zero and a small error in its estimation should be negligible (Vekemans and Hardy 2004). In contrast, the value chosen for D is important, as there can be large differences in effective and census densities (Frankham et al. 2010; Frantz et al. 2010). Census densities of the deer in New Caledonia have been estimated to range from 1 to 20 deer/km2 in rainforests (Le Bel et al. 2001) and to reach > 100 deer/km2 in some favourable habitats (savannah, tropical dry forest; de Garine-Wichatitsky et al. 2009). In general, effective population sizes are approximately 1/10 of census population sizes (Frankham et al. 2010). In order to generate a range of possible values, we estimated σ assuming maximum and minimum effective densities of 100 deer/km2 and 1 deer/km2.
Data analysis: Landscape genetics
Functional connectivity was assessed using ResistanceGA 4.2–8 (Peterman 2018). This R package allows the evaluation of how landscape features influence genetic connectivity by statistically relating the distribution of genetic distance among individuals to landscape characteristics associated with alternative landscape resistance models. It makes use of a machine learning algorithm (a genetic algorithm in R package GA; Scrucca 2013) to optimise resistance surfaces with genetic data, thus avoiding user-specified resistance values. We used log-likelihood as the objective function during the optimisation process which was obtained from linear mixed-effects models fit with pairwise genetic distance as the response variable and pairwise resistance distance as the explanatory variable. Mixed-effects models were fitted using the maximum likelihood population effects (MLPE) parameterisation implemented in R package lme4 (Bates et al. 2015), in order to account for non-independence among pairwise genetic and environmental distances. We assessed the support for optimised resistance surfaces based on Akaike information criteria corrected for small sample size (AICc).
ResistanceGA can optimise categorical and continuous resistance surfaces, as well as multiple resistance surfaces simultaneously. All analyses in this work were based on continuous surfaces. We used the Circuitscape library (Hall et al. 2021) in program Julia v.1.6.7 (Bezanson et al. 2017) to calculate pairwise inter-individual resistance distances. Using the GA.PREP() function, we defined a range of 1–5000 for the resistance values of continuous surfaces to be assessed during optimization. If the difference in AICc (ΔAICc) between two models was > 2 AICc units, the model with the smaller AICc value was considered to be a better fit.
We obtained our spatial data (Table S2) from the geographical information portal of the New Caledonian government (https://georep.nc/). We chose to analyse the impact of eight landscape features: (i) agricultural/open areas: including all agricultural land and open areas with little or no vegetation; (ii) built-up areas: including all artificial habitat, such as urbanised areas (including urban green areas), industrial zones and landfill sites; (iii) dry forest/shrubland: former area of dry forest that nowadays is shrubland consisting mostly of introduced plant species; (iv) forests: including all woodland and tree plantations (excluding the former area of dry forest); (v) mines: all land and infrastructure associated with mining; (vi) roads: all sealed roads; (vii) ultramafic soils: distribution of ultramafic soils. The soil of Grande Terre is composed of either metamorphic and volcano-sedimentary rocks or ultramafic rocks (Isnard et al. 2016). Ultramafic soils have a reduced fertility and the floristic assemblage differs significantly from other substrates (Morat 1993; Isnard et al. 2016). Soils can have an effect on the distribution of rusa deer in New Caledonia (Rouys and Theuerkauf 2003); (viii) water bodies: all water bodies.
We used ArcMap v.10.3 (ERSI Inc.), to generate rasters with 2000 × 2000 m grids. We chose this resolution as a compromise between the scale of the underlying process affecting the relationship between gene flow and landscape structure in a large mobile mammal and computational speed of the analysis. Also, the lack of precise coordinates precluded an analysis using a finer grain. We used two different approaches to parameterise the resistance surfaces corresponding to each feature. First, we calculated the percentage of each feature within each grid cell. In simulation studies, the optimisation of continuous resistance surfaces (rather than categorical surfaces) led to more accurate inference of landscape resistance (Cushman and Landguth 2010; Peterman et al. 2019). Second, we used the gDistance() function in R package rgeos v.0.5–2 (Bivand and Rundel 2019) to calculate the distance from the centroid of each grid cell to the nearest polygon of each respective landscape feature. We considered this approach, as in some instances the suitability of a habitat feature may increase (e.g. distance from road infrastructure) or decrease (e.g. distance from water source) following a certain pattern. Similarly, the avoidance of certain man-made environmental features may increase or decrease depending on the distance from the features in question (e.g. distances from road infrastructure, built-up areas and mining activities; Kimmig et al. 2020).
Furthermore, based on the digital elevation model, we analysed the following features: (ix) elevation: a reduced-resolution version of the digital elevation model; (x) slope: calculated from the reduced-resolution digital elevation model; (xi) roughness: the difference between the maximum and the minimum value of a grid cell of the reduced-resolution digital elevation model and its eight surrounding cells. We used R package raster v.3.1–5 (Hijmans 2020) to aggregate the 100 × 100 m grid cells of the original elevation model to 2000 × 2000 m grids and assigned the median elevation of the smaller grid cells to each new cell. We used the terrain() function of raster to calculate the slope and roughness of each cell (both in radians) using its eight neighbouring cells.
Inter-individual genetic distance metrics are not all equally accurate in model selection in a landscape genetic framework (Beninde et al. 2023). Following Beninde et al. (2023), we therefore aimed to analyse the data using 28 different genetic distance metrics. We considered the following eight metrics: (i) Kc.Lo (referred to as Loiselle’s kinship coefficient Fij above); (ii) Kc.R (Ritland 1996); (iii) Rc.L&R (Lynch and Ritland 1999); (iv) Rc.Li (Li et al. 1993); (v) Rc.Q&G (Queller and Goodnight 1989); (vi) Rc.W (Wang 2002); (vii) Rousset’s â (Rousset 2000); (viii) the proportion of shared alleles DPS (Bowcock et al. 1994). With the exception of DPS, which was estimated using the R package adegenet 2.1.1 (Jombart 2008), all metrics were calculated using Spagedi. We furthermore considered 20 metrics that were derived from a principal component analysis (PCA) or the closely related factorial correspondence analysis (FCA). We used genetix to create an allele count contingency Table (0, 1, or 2) per individual for all alleles in the population. Subsequently, we utilised the R package ade 4 1.7.13 (Dray and Dufour 2007) to conduct FCAs and PCAs (with rescaled allele counts) on the contingency table, determining the position of each individual across axes ranging from 1 to 10. Next, we employed the R package ecodist 2.0.1 (Goslee and Urban 2007) to generate genetic distance matrices based on the Euclidean distance between individual positions on an increasing number of axes (first matrix based on position on axis 1, second matrix based on positions on axes 1 and 2, and so on) up to the first 10 axes. Please refer to Beninde et al. (2023) for further information on the characteristics of all distance metrics.
ResistanceGA is unlikely to perform meaningful optimisations based on a non-significant IBD slopes (B. Peterman, pers. comm.). We therefore first tested for the presence of a significant isolation-by-distance (IBD) pattern when employing each metric in the regression of genetic distance/relatedness metric against the natural logarithm of inter-individual straight-line geographic distances. Again, to reduce bias from the inclusion of pre-dispersal individuals, we performed these tests for adult females (N = 251), all adult males (N = 72) and all adult animals (251 females, 72 males and 33 adults of unknown sex; Fig. 1b). The allele frequencies obtained from the complete data set were used in the separate analyses for males and females. In the case of the FCA- and PCA-based metrics, the position of each individual in the analysis of the complete dataset was used during the separate analyses for males, females and all individuals. We tested for significance by randomisation of spatial location using a custom-written R script (Appendix S1). We only performed the ResistanceGA optimisation for metrics that gave rise to a significant IBD pattern.
For each of the genetic metrics that fulfilled this criterion, we first used the SS_optim() command in ResistanceGA to optimise the resistance of all single environmental predictors (Appendix S2). To check for convergence, we executed each optimisation run twice. We then used the resist.boot() command to perform 1000 iterations of a (pseudo-)bootstrap procedure (Appendix S2). This was done in order to assess the relative support of each optimised resistance surface and the robustness of the model selection results given different sample combinations. In this procedure, individuals and resistance matrices are subsampled without replacement at each iteration, the MLPE model is refitted to different resistance distance matrices and AICc scores are recalculated. We sampled 75% of the observations at each iteration. While we performed two optimisation runs for each predictor, we only used the results of the optimisation with the lower AICc value for bootstrapping.
We then performed multi-surface analyses using the MS_optim() function. For each genetic distance measure, we performed those analyses for all combinations of predictors that explained gene flow patterns better than the distance-only model (difference in AICc > 2) in the initial single-predictor analysis. However, when both versions of an environmental feature (the percentage of each feature within each grid cell and the distance from the centroid of each grid cell to the nearest polygon) explained gene flow better than distance, we only included the version with the lower AICc value in the multi-surface analysis. To check for convergence, we executed each optimisation run twice. We then used the resist.boot() function to perform a bootstrap procedure on the original better-than-distance models and the corresponding multi-surface models (1000 iterations, 75% of the observations sampled at each iteration). We only used the results of the optimisation run with the lowest AICc value for bootstrapping.
Results
When analysing the complete dataset, five loci deviated from HWE after correcting for multiple test (BM1818, ETH225, OarCP26, OarFCB304, T156; P < 0.0156). However, our loci did not systematically deviate from HWE when analysing geographically coherent subsamples (arbitrarily based on political boundaries). When subdividing the dataset into municipalities, all the loci were in HWE in nine of these 13 pre-defined populations and no locus deviated from HWE in more than three municipalities (Table S2). Furthermore, only five different pairs of loci were in linkage disequilibrium across all 13 municipalities after correcting for multiple tests. We did not find any evidence for the presence of null alleles. All loci were thus retained for further analysis.
Structure did not provide evidence for population genetic structure as the highest log-likelihood values were obtained for K = 1 (Fig. 2). Similarly, when performing spatial clustering of individuals, BAPS inferred a probability of p(S) = 1 for the presence of one genetic population. The loci were characterised by an average of 3.4 alleles (SD 1.6; range 2–7), an average observed heterozygosity of Ho = 0.526 (SD 0.131; range 0.327–0.709) and an average expected heterozygosity of He = 0.512 (SD 0.129; 0.337–0.750; Table 1).
Based on Loiselle’s kinship coefficient (Kc.Lo), we obtained a significant IBD pattern when considering all adults (slope = − 0.0022; s.e.: 0.0004; P < 0.001). However, when focusing on the different sexes, the IBD pattern was significant for females (slope = − 0.0017; s.e.: 0.0005; P < 0.001), but not for males (slope = 0.0021; s.e.: 0.0012; P = 0.228). The average axial mother-daughter distance (σ) was estimated to range between 0.62 km (effective density 100 deer/km2) and 6.17 km (1 deer/km2). The average pairwise distance between females was 89.52 km (SD 66.28) and 67.27 km (SD 64.07) between males, but males had larger maximal pairwise geographic distances than females (males, 488.7 km; females, 332.0; Fig. S1).
Aside from Loiselle’s kinship coefficient (Kc.Lo), only the Kc.R, Rc.L&R and Rc.Q&G metrics gave rise to a significant IBD pattern both when considering adult females or all adults (Table S3). No other metric gave rise to a significant IBD pattern in any of the three categories considered. When optimising landscape resistance surfaces based on adult females, no single environmental factor explained gene flow patterns better than the Euclidean distance between pairs of animals when using the Kc.Lo, Kc.R and Rc.L&R metrics for single-surface optimisation (Tables S4–S6). In the case of the Rc.Q&G metric, two environmental features explained gene flow patterns better than the distance-only model (Table S7). However, the corresponding multi-feature model did not outperformed the best single-feature model (ΔAICc < 2; Table 2), but the best single-feature model distance to shrubland performed extremely poorly overall (mR2 = 0.0038).
When considering all adults, no single environmental factor better explained gene flow patterns than the Euclidean distance between pairs of animals when using the Kc.Lo and Kc.R metrics for single-surface optimisation (Tables S4–S5). In contrast, five and seven environmental features explained gene flow patterns better than the distance-only model when using the Rc.Q&G and Rc.L&R metrics, respectively (Tables S6–S7). In the case of both metrics, no multi-feature model outperformed the best single-feature model (ΔAICc < 2; Tables 2 and 3). In both cases, the best single-feature model (Rc.Q&G: distance to shrubland; Rc.L&R: distance to roads) performed extremely poorly overall (Rc.Q&G: mR2 = 0.0043; Rc.L&R: mR2 = 0.0018).
Discussion
We analysed the population and landscape genetic structure of rusa deer in New Caledonia to identify natural barriers and understand landscape resistance in order to generate basic ecological knowledge that may help optimise management strategies. We did not find any evidence for the presence of population genetic structure, providing evidence for the population being interconnected across the whole of the Grande Terre Island and for the absence of significant movement barriers.
Our results are in line with the landscape only having a very limited effect on deer dispersal. The Bayesian clustering algorithms did not provide evidence for a genetic discontinuity in the study area. While the presence of an IBD pattern can also be indicative of the presence of genetic discontinuities (Guillot et al. 2009), we did not find evidence for IBD when focusing on males. Landscape resistance only had a very minor, if any, role in the dispersal of rusa deer at the scale of the Grande Terre Island.
Depending on the chosen metric, either no single environmental factor better explained gene flow patterns than the Euclidean distance or the best-supported models had extremely low power (mR2 ≤ 0.0043) to accurately predict the distribution of genetic distances among individuals based on landscape resistance models. Other individual-based landscape genetic studies have reported mR2 > 0.3 for the best ResistanceGA landscape resistance model (e.g. Kimmig et al. 2020; Bauder et al. 2021).
Following Vekemans and Hardy (2004), we chose Loiselle’s kinship coefficient Fij for estimating the extent of the fine-scale genetic structure in the dataset and to derive the axial mother-daughter distance. Our results showed a significant difference in patterns of fine-scale genetic structure between males and females. A similar result was obtained when comparing the IBD patterns of female and males using the Kc.R, Rc.L&R and Rc.Q&G metrics. These IBD patterns obtained using these four metrics thus suggested that the dispersal of rusa deer in New Caledonia was male-biased, a result that has also been observed in red deer (Frantz et al. 2008). The absence of IBD implies in principle that male dispersal distances were large compared to the size of the study area (Guillot et al. 2009). While telemetry studies reported rusa deer to have home ranges of a few square kilometres (Moriarty 2004; Spaggiari and de Garine-Wichatitsky 2006; Amos et al. 2022), there is also evidence for sub-adult males dispersing from high- to low-density deer areas in a study area in Australia (Moriarty 2004). It should be mentioned, however, that, in order to compare IBD patterns in a meaningful way, datasets should have similar distributions of pairwise geographic distances (Frantz et al. 2010). Here, maximum pairwise geographic distances were larger in males than in females, and the absence of IBD in males was thus not due to the analysis being performed over a smaller spatial extent. Nevertheless, we cannot completely rule out the possibility that the absence of an IBD pattern in males is an artefact of the sampling distribution.
Depending on the effective density, the average female dispersal distance was estimated to range between 0.62 and 6.17 km. In favourable habitats, where effective densities are bound to be higher than 1 deer/km2, female dispersal distances are thus likely to be limited to a few kilometres on average. These distances must be interpreted with caution as effective density estimates were approximate and will vary by habitat type. In addition, in contrast to males (Moriarty 2004), females possibly remain with their mothers at least for the first 2 years of their lives, as they do in red deer (Bützler 1986) and fallow deer (Dama dama; Heidemann 1986). Some females with permanent incisors may thus not have dispersed yet, leading to an overestimate of the strength of the IBD slope and an underestimate of dispersal distances. Despite these caveats, female dispersal distances are likely to be small in relation to the size of the study area.
A potential problem with our analyses is that the initial introduction and range expansion may be too recent for the effects of mutation and drift to be at equilibrium (Fitzpatrick et al. 2012). After their introduction 150 years ago, the deer underwent a rapid expansion, colonising the whole island by the 1940s at the latest. Landguth et al. (2010) have performed simulations to assess the time lag between creating a physical barrier and detecting a genetic signal. They showed that an individual-based landscape resistance approach using DPS as a genetic distance measure generally detected a new barrier 15 generations after its establishment. Furthermore, population genetic systems that are mainly characterised by drift or that preserve genetic signals of historical connectivity should not be characterised by isolation-by-distance patterns (Hutchison and Templeton 1999). The fact that we observe an isolation-by-distance pattern in females, but not males, is thus likely to be a genuine result indicative of female philopatry. Therefore, considering that we detected only a minor influence of the landscape on dispersal pattern and that male dispersal was large relative to the site of the study area, we believe that the absence of population genetic structure was a biologically meaningful result.
Conservation implications
One of the key measures of the management plan for rusa deer in New Caledonia has been the identification of ten priority areas for deer control operations. Large-scale control operations, including helicopter-based hunts, have been performed in three of these areas (Anonymous 2022). Our results showed that the deer formed a single genetic population on the main New Caledonian island and had large dispersal distances, with gene flow barely affected by landscape features. It is uncertain whether a sufficiently large proportion of deer is removed to effectively reduce population sizes within the management areas (Tron and Barrière 2016). In any case, our results suggest that the removal of deer from priority areas is likely to create a population sink, with natural dispersal offsetting the removal operations.
The obvious solution to controlling an interconnected population is to reduce deer population size to sustainable levels across the whole of Grande Terre. Even if large-scale helicopter-based operations could feasibly reduce island-wide population densities, this approach is likely prohibitively expensive (Tron and Barrière 2016) and would need to be repeated regularly. Moreover, despite their serious ecological impact, the deer are of major cultural and economic significance. Venison constitutes a key source of animal protein for local communities and hunting plays an important role in the life of many New Caledonians (De Garine-Wichatitsky et al. 2004). It is thus far from certain that large-scale control, even if feasible, would gain societal acceptance (De Garine-Wichatitsky et al. 2004; Tron and Barrière 2016).
Availability of data and material
An XLSX file containing the geo-referenced microsatellite genotypes is available on Figshare. https://doi.org/10.6084/m9.figshare.22651462.
Code availability
Appendices 1–3 in Supplementary Materials.
References
Amos M, Pople A, Brennan M, Sheil D, Kimber M, Cathcart A (2022) Home ranges of rusa deer (Cervus timorensis) in a subtropical peri-urban environment in South East Queensland. Aust Mammal 45:116–120. https://doi.org/10.1071/AM21052
Anderson EC, Dunham KK (2008) The influence of family groups on inferences made with the program Structure. Mol Ecol 8:1219–1229. https://doi.org/10.1111/j.1755-0998.2008.02355.x
Anonymous (2022) Réalisation d’opérations de lutte contre les ongulés envahissants. Projet Régional Océanien des Territoires pour la Gestion durable des Ecosystèmes. https://protege.spc.int/sites/default/files/documents/22_09_PROTEGE-FicheSuivi-EspecesEnvahissantes-13B.1_BD.pdf. Accessed 18 April 2023
Aronson J, Vallaur D, Jaffré T, Lowry P II (2005) Restoring dry tropical forest. In: Mansourian S, Vallauri D, Dudley N (eds) Restoring forests and their functions in landscapes, beyond planting trees. Springer, New York, pp 285–290
Barendse W, Armitage SM, Kossarek L et al (1994) A genetic linkage map of the bovine genome. Nat Genet 6:227–235. https://doi.org/10.1038/ng0394-227
Barrière P, Fort C (2021) Rusa timorensis (de Blainville, 1822). In: Savouré-Soubelet A, Arthur CP, Aulagnier S, Body G, Callou C, Haffner P, Marchandeau S, Moutou F, Saint-Andrieux C (eds) Atlas des mammifères sauvages de France, vol 2. Ongulés et Lagomorphes. Muséum National d’Histoire Naturelle, Paris, pp 96–101
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Bauder JM, Peterman WE, Spear SF, Jenkins CL, Whiteley AR, McGarigal K (2021) Multiscale assessment of functional connectivity: landscape genetics of eastern indigo snakes in an anthropogenically fragmented landscape in central Florida. Mol Ecol 30:3422–3438. https://doi.org/10.1111/mec.15979
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier.
Beninde J, Wittische J, Frantz AC (2023) Quantifying uncertainty in inferences of landscape genetic resistance due to choice of individual-based genetic distance metric. Mol Ecol Resour. https://doi.org/10.1111/1755-0998.13831
Bezanson J, Edelman A, Karpinski S, Shah VB (2017) Julia: a fresh approach to numerical computing. SIAM Rev 59:65–98. https://doi.org/10.1137/141000671
Bianchi M, Hurlin JC, Lebel S, Chardonnet P (1997) Observations on the eruption of the permanent incisor teeth of farmed Javan rusa deer (Cervus timorensis russa) in New Caledonia. N Z Vet J 45:167–168. https://doi.org/10.1080/00480169.1997.36018
Bishop MD, Kappe SM, Keele JW et al (1994) A genetic linkage map for cattle. Genetics 136:619–639. https://doi.org/10.1093/genetics/136.2.619
Bivand R, Rundel C (2019) rgeos: Interface to Geometry Engine - Open Source (‘GEOS’). R package version 0.5–2. https://CRAN.R-project.org/package=rgeos. Accessed 21 Sept 2022
Bonnet A, Thévenon S, Maudet F, Maillard JC (2002) Efficiency of semi-automated fluorescent multiplex PCRs with 11 microsatellite markers for genetic studies of deer populations. Anim Genet 33:343–350. https://doi.org/10.1046/j.1365-2052.2002.00873.x
Bourgain C, Abney M, Schneider D, Ober C, McPeek MS (2004) Testing for Hardy-Weinberg equilibrium in samples with related individuals. Genetics 168:2349–2361. https://doi.org/10.1534/genetics.104.031617
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455. https://doi.org/10.1038/368455a0
Buchanan FC, Crawford AM (1993) Ovine microsatellites at the OarFCB11, OarFCB128, OarFCB193, OarFCB266 and OarFCB304 loci. Anim Genet 24:145. https://doi.org/10.1111/j.1365-2052.1993.tb00269.x
Buchanan FC, Galoway SM, Crawford AM (1994) Ovine micro-satellites at the OarFCB5, OarFCB19, OarFCB20, OarFCB48, OarFCB129, and OarFCB226 loci. Anim Genet 25:60. https://doi.org/10.1111/j.1365-2052.1994.tb00461.x
Bützler W (1986) Cervus elaphus (Linnaeus, 1758) – Rothirsch. In: Niethammer J, Krapp F (eds) Handbuch der Säugetiere Europas: Paarhufer. Aula Verlag, Wiesbaden, pp 107–139
Chatelier J (2007) La révision toponymique (et cartographique) en Nouvelle-Calédonie (1983–1993). J Soc Océan 125:295–310. https://doi.org/10.4000/jso.1020
Coltman DW, Pilkington JG, Pemberton JM (2003) Fine-scale genetic structure in a free-living ungulate population. Mol Ecol 12:733–742. https://doi.org/10.1046/j.1365-294X.2003.01762.x
Comer CE, Kilgo JC, D’Angelo GJ, Glenn TC, Miller KV (2005) Fine-scale genetic structure and social organization in female white-tailed deer. J Wildl Manage 69:332–344. https://doi.org/10.2193/0022-541X(2005)069%3c0332:FGSASO%3e2.0.CO;2
Corander J, Siren J, Arjas E (2008) Bayesian spatial modeling of genetic population structure. Comput Stat 23:111–129. https://doi.org/10.1007/s00180-007-0072-x
Courchamp F, Chapuis JL, Pascal M (2003) Mammal invaders on islands: impact, control and control impact. Biol Rev 78:347–383. https://doi.org/10.1017/s1464793102006061
Cushman SA, Landguth EL (2010) Scale dependent inference in landscape genetics. Landscape Ecol 25:967–979. https://doi.org/10.1007/s10980-010-9467-0
De Garine-Wichatitsky M, Duncan P, Labbe A, Supriw D, Chardonnet P, Maillard D (2003) A review of the diet of rusa deer Cervus timorensis russa in New Caledonia: are the endemic plants defenceless against this introduced, eruptive ruminant? Pac Conserv Biol 9:136–143. https://doi.org/10.1071/pc030136
De Garine-Wichatitsky M, Chardonnet P, de Garine I (2004) Management of introduced game species in New Caledonia: reconciling biodiversity conservation and resource use? Game Wildl Sci 21:697–706
De Garine-Wichatitsky M, Soubeyran Y, Maillard D, Duncan P (2005) The diets of introduced rusa deer (Cervus timorensis russa) in a native sclerophyll forest and a native rainforest of New Caledonia. N Z J Zool 32:117–126. https://doi.org/10.1080/03014223.2005.9518403
De Garine-Wichatitsky M, de Meeûs T, Chevillon C, Berthier D, Barré N, Thévenon S, Maillard JC (2009) Population genetic structure of wild and farmed rusa deer (Cervus timorensis russa) in New-Caledonia inferred from polymorphic microsatellite loci. Genetica 137:313–323. https://doi.org/10.1007/s10709-009-9395-6
de Wit LA, Zilliacus KM, Quadri P et al (2020) Invasive vertebrate eradications on islands as a tool for implementing global Sustainable Development Goals. Environ Conserv 47:139–148. https://doi.org/10.1017/s0376892920000211
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Dumas P (2013) Anthropization of New Caledonia’s southwestern coastland: a satellite remote-sensing analysis of coastland environment changes. In: Larrue S (ed) Biodiversity and societies in the Pacific Islands. Presses Universitaires de Provence/Australian National University, Aix-en-Provence, pp 113–129
Ede AJ, Pierson CA, Crawford AM (1995) Ovine microsatellites at the OarCP9, OarCP16, OarCP20, OarCP21, OarCP23, and OarCP26 loci. Anim Genet 25:129–130. https://doi.org/10.1111/j.1365-2052.1995.tb02655.x
Fitzpatrick BM, Fordyce JA, Niemiller ML, Reynolds RG (2012) What can DNA tell us about biological invasions? Biol Invasions 14:245–253. https://doi.org/10.1007/s10530-011-0064-1
Fotsing JM, Dumas P (2021) The physical and human geography of the New Caledonian archipelago. In: Gravelat E (ed) Understanding New Caledonia. University Press of New Caledonia, Nouméa, pp 19–36
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to conservation genetics, 2nd edn. Cambridge University Press, Cambridge
Frantz AC, Hamann JL, Klein F (2008) Fine-scale genetic structure of red deer (Cervus elaphus) in a French temperate forest. Eur J Wildl Res 54:44–52. https://doi.org/10.1007/s10344-007-0107-1
Frantz AC, Cellina S, Krier A, Schley L, Burke T (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol 46:493–505. https://doi.org/10.1111/j.1365-2664.2008.01606.x
Frantz AC, Do Linh San E, Pope LC, Burke T (2010) Using genetic methods to investigate dispersal in two badger (Meles meles) populations with different ecological characteristics. Heredity 104:493–501. https://doi.org/10.1038/hdy.2009.136
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19. https://doi.org/10.18637/jss.v022.i07
Guillot G, Leblois R, Coulon A, Frantz AC (2009) Statistical methods in spatial genetics. Mol Ecol 18:4734–4756. https://doi.org/10.1111/j.1365-294x.2009.04410.x
Hall KR, Anantharaman R, Landau VA et al (2021) Circuitscape in Julia: empowering dynamic approaches to connectivity assessment. Land 10:301. https://doi.org/10.3390/land10030301
Hardy OJ, Vekemans X (2002) Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8286.2002.00305.x
Heidemann G (1986) Cervus dama (Linnaeus, 1758) – Damhirsch. In: Niethammer J, Krapp F (eds) Handbuch der Säugetiere Europas: Paarhufer. Aula Verlag, Wiesbaden, pp 140–158
Hijmans RJ (2020) raster: Geographic Data Analysis and Modeling. R package version 3.1–5. https://CRAN.R-project.org/package=raster. Accessed 21 Sept 2022
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914. https://doi.org/10.2307/2640449
Isnard S, L’huillier L, Rigault F, Jaffré T (2016) How did the ultramafic soils shape the flora of the New Caledonian hotspot? Plant Soil 403:53–76. https://doi.org/10.1007/s11104-016-2910-5
Jaffré T, Rigault F, Dagostini G, Fambart-Tinel J, Wulff A, Munzinger J (2009) Input of the different vegetation units to the richness and endemicity of the New Caledonian flora. In: Mery P (ed) Proceedings of the 11th Pacific Science Inter-Congress, Tahiti, French Polynesia, 2–6 March, 2009. Pacific Science Association, Honolulu
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jones KC, Levine KF, Banks JD (2002) Characterization of 11 polymorphic tetranucleotide microsatellites for forensic applications in California elk (Cervus elaphus canadensis). Mol Ecol Notes 2:425–427. https://doi.org/10.1046/j.1471-8286.2002.00264.x
Kimmig SE, Beninde J, Brandt M et al (2020) Beyond the landscape: resistance modelling infers physical and behavioural gene flow barriers to a mobile carnivore across a metropolitan area. Mol Ecol 26:466–484. https://doi.org/10.1111/mec.15345
Kuehn R, Schroeder W, Pirchner F, Rottmann O (2003) Genetic diversity, gene flow and drift in Bavarian red deer population (Cervus elaphus). Conserv Genet 4:157–166. https://doi.org/10.1023/A:1023394707884
Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191. https://doi.org/10.1111/j.1365-294x.2010.04808.x
Le Bel S, Sarrailh JM, Brescia F, Cornu A (2001) Présence du cerf rusa dans le massif de l’Aoupinié en Nouvelle-Calédonie et impact sur le reboisement en kaoris. Bois Et Forêts Des Trop 269:5–18
Li CC, Weeks DE, Chakravarti A (1993) Similarity of DNA fingerprints due to chance and relatedness. Hum Hered 43:45–52. https://doi.org/10.1159/000154113
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understorey shrub Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425. https://doi.org/10.1002/j.1537-2197.1995.tb12679.x
Losfeld G, L’Huillier L, Fogliani B, Jaffré T, Grison C (2015) Mining in New Caledonia: environmental stakes and restoration opportunities. Environ Sci Pollut Res 22:5592–5607. https://doi.org/10.1007/s11356-014-3358-x
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766. https://doi.org/10.1093/genetics/152.4.1753
Mansourian S, Géraux H, Do Khac E, Vallauri D (2018) Lessons learnt from 17 years of restoration in New Caledonia’s dry tropical forest. WWF, France
McRae BH (2006) Isolation by resistance. Evolution 60:1551–1561. https://doi.org/10.1111/j.0014-3820.2006.tb00500.x
Mezzelani A, Zhang Y, Redaelli L et al (1995) Chromosomal localization and molecular characterization of 53 cosmid-derived bovine microsatellites. Mamm Genome 6:629–635. https://doi.org/10.1007/bf00352370
Miller SA, Dykes DD, Polesky HF (1988) A simple salting-out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. https://doi.org/10.1093/nar/16.3.1215
Mommens G, Coppieters W, Van de Weghe A, Van Zeveren A, Bouquet Y (1994) Dinucleotide repeat polymorphism at the bovine MM12E6 and MM8D3 loci. Anim Genet 25:368. https://doi.org/10.1111/j.1365-2052.1994.tb00381.x
Moore SS, Byrne K, Berger KT, Barendse W, McCarthy F, Womack JE, Hetzel DJS (1994) Characterization of 65 bovine microsatellites. Mamm Genome 5:84–90. https://doi.org/10.1111/j.1365-2052.1994.tb00381.x
Morat P (1993) Our knowledge of the flora of New Caledonia: endemism and diversity in relation to vegetation types and substrates. Biodivers Lett 1:71–81. https://doi.org/10.2307/2999750
Morat P, Jaffré T, Tronchet F, Munzinger J, Pillon Y, Veillon JM, Chalopin M (2012) The taxonomic reference base Florical and characteristics of the native vascular flora of New Caledonia. Adansonia 34:177–219. https://doi.org/10.5252/a2012n2a1
Moriarty AJ (2004) Ecology and environmental impact of Javan rusa deer (Cervus timorensis russa) in the Royal National Park. PhD Thesis, University of Western Sydney
Munzinger J, Morat P, Jaffré T et al. (2022) FLORICAL: checklist of the vascular indigenous flora of New Caledonia. http://publish.plantnet-project.org/project/florical. Accessed 21 Sept 2022
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858. https://doi.org/10.1038/35002501
Pascal M, Richer de Forges B, Le Guyader H, Simberloff D (2008) Mining and other threats to the New Caledonia biodiversity hotspot. Conserv Biol 22:498–499. https://doi.org/10.1111/j.1523-1739.2008.00889.x
Pérez-Espona S, Pérez-Barbería FJ, McLeod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17:981–996. https://doi.org/10.1111/j.1365-294x.2007.03629.x
Peterman WE (2018) ResistanceGA: an R package for the optimisation of resistance surfaces using genetic algorithms. Methods Ecol Evol 9:1638–1647. https://doi.org/10.1111/2041-210X.12984
Peterman WE, Winiarski KJ, Moore CE, da Silva CC, Gilbert AL, Spear SF (2019) A comparison of popular approaches to optimize landscape resistance surfaces. Landsc Ecol 34:2197–2208. https://doi.org/10.1007/s10980-019-00870-3
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275. https://doi.org/10.1111/j.1558-5646.1989.tb04226.x
Raymond M, Rousset F (1995) GENEPOP (version 1.2): a population genetics software for exact tests and ecumenicism. J Hered 86:248–249. https://doi.org/10.1093/oxfordjournals.jhered.a111573
Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67:175–185. https://doi.org/10.1017/S0016672300033620
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62. https://doi.org/10.1046/j.1420-9101.2000.00137.x
Rouys S, Theuerkauf J (2003) Factors determining the distribution of introduced mammals in nature reserves of the southern province, New Caledonia. Wildl Res 30:187–191. https://doi.org/10.1071/WR01116
Sax DF, Gaines SD (2008) Species invasions and extinction: the future of native biodiversity on islands. Proc Natl Acad Sci USA 105:11490–11497. https://doi.org/10.1073/pnas.0802290105
Scrucca L (2013) GA: a package for genetic algorithms in R. J Stat Softw 53:1–37. https://doi.org/10.18637/jss.v053.i04
Spaggiari J, De Garine-Wichatitsky M (2006) Home range and habitat use of introduced rusa deer (Cervus timorensis russa) in a mosaic of savannah and native sclerophyll forest in New Caledonia. N Z J Zool 33:175–183. https://doi.org/10.1080/03014223.2006.9518442
Steffen P, Eggen A, Dietz AB, Womack JE, Stranzinger G, Fries R (1993) Isolation and mapping of polymorphic microsatellites in cattle. Anim Genet 24:121–124. https://doi.org/10.1111/j.1365-2052.1993.tb00252.x
Tramier CMC, Genthon P, Delvienne QRCP et al (2021) Hydrological regimes in a tropical valley of New Caledonia (SW Pacific): impacts of wildfires and invasive fauna. Hydrol Process 35:e14071. https://doi.org/10.1002/hyp.14071
Tron F, Barrière P (2016) Eléments de cadrage pour une stratégie de régulation des cerfs en Nouvelle-Calédonie: zones prioritaires, vision, objectifs et ressources nécessaires. Conservation International et Conservatoire d’Espaces Naturels de Nouvelle-Calédonie.
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. https://doi.org/10.1046/j.1365-294x.2004.02076.x
Verhoeven KJF, Simonsen KL, McIntyre LM (2005) Implementing false discovery rate control: increasing your power. Oikos 108:643–647. https://doi.org/10.1111/j.0030-1299.2005.13727.x
Wang J (2002) An estimator for pairwise relatedness using molecular markers. Genetics 160:1203–1215. https://doi.org/10.1093/genetics/160.3.1203
Woodroffe R, Thirgood S, Rabinowitz A (2005) The impact of human–wildlife conflict on natural systems. In: Woodroffe R, Thirgood S, Rabinowitz A (eds) People and wildlife: conflict or coexistence. Cambridge University Press, Cambridge
Wulff AS, Hollingsworth PM, Ahrends A, Jaffré T, Veillon JM, L’Huillier L, Fogliani B (2013) Conservation priorities in a biodiversity hotspot: analysis of narrow endemic plant species in New Caledonia. PLoS ONE 8:e73371. https://doi.org/10.1371/journal.pone.0073371
Acknowledgements
We would like to thank the numerous individuals who contributed their valuable assistance in the field. Mr. Philippe Severian, head of the “Direction du Développement Rural—Province Sud”, and Mr. Lionnel Brinon, head of the “Agence pour la prévention et l’indemnisation des calamités agricoles ou naturelles (Apican)” and President of the “Agence Rurale”, obtained the financial resources necessary for the realisation of the fieldwork. We would like to thank both Neo-Caledonian provinces, the “Institut Agronomique Néo-Calédonien” and the “Agence Rurale” for their support of the project. Some of the samples were provided by the GEE of the “Conservatoire des Espaces Naturels”. We would like to thank Anna Schleimer, Julian Wittische and two anonymous reviewers for comments on earlier versions of the manuscript.
Funding
This work was conducted in the framework of the “Dental Microwear Texture Analyses (DMTA) Rusa” scientific project funded by the Agence pour l’Indemnisation des Calamités Agricoles ou Naturelles (APICAN)/Agence rurale (P.I.: E. Berlioz, conventions: 4922/407/APICAN/18; AR/2020-03-345) and by the postdoctoral L ‘Oréal-UNESCO For Women in Science Grant obtained by E. Berlioz in 2019. The laboratory work was funded by an internal grant from the National Natural History Museum Luxembourg.
Author information
Authors and Affiliations
Contributions
Alain Frantz, Marc Colyn and Emilie Berlioz conceived and organised the project; Emilie Berlioz collected all the samples in the field, Amanda Luttringer and Christos Kazilas performed the laboratory work, Alain Frantz analysed the data. Alain Frantz wrote the manuscript with contributions from Emilie Berlioz. All the authors commented on the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Tissue samples were collected in accordance with the Nagoya Protocol (collection ID: 20200331102055171).
Consent to participate
All authors gave consent to participate.
Consent for publication
All authors gave consent for publication.
Competing interests
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Frantz, A.C., Luttringer, A., Colyn, M. et al. Landscape structure does not hinder the dispersal of an invasive herbivorous mammal in the New Caledonian biodiversity hotspot. Eur J Wildl Res 70, 6 (2024). https://doi.org/10.1007/s10344-023-01757-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-023-01757-0