Abstract
Background
Endoscopic ultrasonography (EUS) is useful for the differential diagnosis of subepithelial lesions (SELs); however, not all of them are easy to distinguish. Gastrointestinal stromal tumors (GISTs) are the commonest SELs, are considered potentially malignant, and differentiating them from benign SELs is important. Artificial intelligence (AI) using deep learning has developed remarkably in the medical field. This study aimed to investigate the efficacy of an AI system for classifying SELs on EUS images.
Methods
EUS images of pathologically confirmed upper gastrointestinal SELs (GIST, leiomyoma, schwannoma, neuroendocrine tumor [NET], and ectopic pancreas) were collected from 12 hospitals. These images were divided into development and test datasets in the ratio of 4:1 using random sampling; the development dataset was divided into training and validation datasets. The same test dataset was diagnosed by two experts and two non-experts.
Results
A total of 16,110 images were collected from 631 cases for the development and test datasets. The accuracy of the AI system for the five-category classification (GIST, leiomyoma, schwannoma, NET, and ectopic pancreas) was 86.1%, which was significantly higher than that of all endoscopists. The sensitivity, specificity, and accuracy of the AI system for differentiating GISTs from non-GISTs were 98.8%, 67.6%, and 89.3%, respectively. Its sensitivity and accuracy were significantly higher than those of all the endoscopists.
Conclusion
The AI system, classifying SELs, showed higher diagnostic performance than that of the experts and may assist in improving the diagnosis of SELs in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The detection rate of subepithelial lesions (SELs) of the upper gastrointestinal tract increases with an increase in the number of upper endoscopies [1, 2]. In the diagnosis of upper gastrointestinal SELs, observation of the originating layer, margins, echogenicity, and detailed morphology of the lesions using endoscopic ultrasonography (EUS) is useful for differential diagnosis; however, not all of them are easy to distinguish [3, 4]. Gastrointestinal stromal tumors (GISTs) are the most common SELs in the gastrointestinal tract and are considered potentially malignant. It is important to differentiate GISTs from benign SELs [5, 6]. Gastrointestinal schwannomas are rare, generally benign, and slow-growing tumors with excellent long-term prognoses after surgical resection [7,8,9]. However, preoperative differentiation of schwannomas from GISTs with different prognoses is difficult, and in the absence of a definite preoperative pathological diagnosis, surgical resection is considered the gold standard treatment [9, 10].
Previous studies have reported the accuracy of differential diagnosis of SELs by EUS ranging from 45.5 to 66.7% [11,12,13]. Because of the insufficient accuracy of differential diagnosis of SELs by EUS, histopathology-based methods, such as EUS-guided fine-needle aspiration biopsy (EUS-FNAB) and mucosal cutting biopsy, are generally recommended to confirm the diagnosis [14,15,16]. However, these methods are invasive and their efficacy depends on the skill of the physician. Moreover, it can be difficult to obtain sufficiently large samples for immunohistochemical analysis using EUS-FNAB [14, 17]. Mucosal cutting biopsy is an invasive method with risks of intraoperative bleeding and perforation which may cause tumor cell seeding, and the sampling of SELs with extramural growth form is difficult [18]. Previous studies reported that the accuracy of EUS-FNAB in SELs ranged widely from 62.0 to 93.4% and was low for small lesions < 2 cm in size, while that of mucosal cutting biopsy ranged from 85.2 to 94.3% [16,17,18,19,20,21]. Due to the risks involved and poor accuracy of invasive biopsy techniques, more accurate and non-invasive diagnostic methods are desired.
Recently, artificial intelligence (AI) using deep learning by convolutional neural networks (CNNs) [22] has developed remarkably in the medical field [23, 24]. It has also been applied to the field of endoscopic diagnosis of esophageal cancer [25], gastric cancer [26], colorectal polyp [27], and EUS diagnosis of pancreatic disease [28]. Previous studies applying AI-based diagnostic systems to EUS images of SELs reported diagnostic yields ranging from 79.2 to 90.0% [29, 30]. However, in these studies, the number of patients included in the training and test datasets was up to 300, and data were collected from a single institution or a small number of institutions (less than four). In addition, the AI-based diagnostic systems in these studies were only applied to binary classification and were not practical enough. AI-based diagnosis of SELs on EUS images based on larger, multicenter data may have higher generalizability and practical applicability in differential diagnosis than previous studies. This study aimed to develop a new AI system for multi-category classifications of common types of SELs (GIST, leiomyoma, schwannoma, neuroendocrine tumor [NET], and ectopic pancreas) on EUS images based on large multicenter data, and to investigate its efficacy.
Methods
Study sample
All EUS images were retrospectively obtained from the Nagoya University Hospital (NUH) along with 11 other Japanese hospitals between January 2005 and December 2020. This study included upper gastrointestinal SEL cases (GIST, leiomyoma, schwannoma, NET, and ectopic pancreas) pathologically confirmed by surgical or endoscopic resection, EUS-FNAB, or other biopsy techniques, and ectopic pancreas cases without pathological diagnoses, which were diagnosed by board-certified specialists at the Japan Gastroenterological Endoscopy Society, based on imaging findings of both conventional endoscopic and EUS examinations. When available, pathological diagnosis based on surgically or endoscopically resected specimens was considered as the reference standard. For SELs in which surgical or endoscopic resection was not performed, immunohistochemical analysis results of EUS-FNAB or other biopsy technique specimens and/or the results of periodic conventional endoscopic and EUS follow-up examinations at intervals of more than six months were considered as alternative reference standards. EUS images were retrieved in the JPEG format. The exclusion criteria were poor quality images resulting due to air, blurring, out of focus, or artifacts.
This study was approved by the Ethics Committees of the NUH (No. 2018-0435-3, date: September 9, 2020) and those of 11 other Japanese hospitals, and was conducted in accordance with the Declaration of Helsinki. The written consent was waived by the ethics committees because the study had a retrospective design.
EUS procedure
EUS was performed using conventional echoendoscopes (GF-UCT240, GF-UE260-AL5, GF-UCT260, TGF-UC260J, or GF-UM2000: Olympus Corporation, Tokyo, Japan; EG-530UR, EG-580UR, or EG-580UT: Fujifilm Corporation, Tokyo, Japan; EG-3830UT or EG-3870UTK: Pentax Lifecare Division, Hoya Co, Ltd, Tokyo, Japan) at 5–20 MHz or mini-probes (UM-2R, frequency 12 MHz, UM-3R, frequency 20 MHz, or UM-DP20-25R, frequency 20 MHz: Olympus Corporation; P-2726–12, frequency 12 MHz or P-2726–20, frequency 20 MHz: Fujifilm Corporation) and ultrasound systems (EU-ME1 or EU-ME2: Olympus Corporation; VP-4450HD, SU-1, or SP900: Fujifilm Corporation; ARIETTA 850, Prosound F75, or Prosound SSD α-10: Hitachi Aloka Medical, Tokyo, Japan). The lesion was scanned after removing mucus and foam and filling the upper gastrointestinal tract with degassed water. The frequency was set high (12 or 20 MHz) for the observation of the originating layer of the lesion and was changed to a lower one (5–7.5 MHz) when the entire image could not be obtained.
Building development and test datasets
All pathologically confirmed EUS SEL images were classified into five categories: GIST, leiomyoma, schwannoma, NET, and ectopic pancreas. In addition, the images were classified into two categories from two perspectives: based on prognosis as GIST or non-GIST and based on treatment strategy as GIST/schwannoma (mesenchymal tumors for which surgical resection was recommended) or other SELs.
In this study, the entire dataset of pathologically confirmed SELs was divided into development and test datasets, which were mutually exclusive, in the ratio of 4:1 using random sampling based on cases, rather than images. Similarly, the development dataset was divided into training and validation datasets in the ratio of 4:1. The dataset of ectopic pancreases without pathological diagnoses was used as an unlabeled dataset for semi-supervised learning [31].
All EUS images of SELs were trimmed to equally sized squares.
Developing the AI system
PyTorch (https://pytorch.org/) was used for the deep learning algorithm, and the original deep learning algorithm based on the EfficientNetV2-L algorithm [32] was used. All CNN layers were fine-tuned from the weights of the pre-trained models with ImageNet21k [33]. In this study, pathologically confirmed EUS SEL images were used as the input information. The data were labeled according to the pathological results in the manner in which SEL was defined (leiomyoma, 0; GIST, 1; schwannoma, 2; NET, 3; ectopic pancreas, 4). All EUS images were trimmed to equally sized squares (224 × 224 pixels) to fit the input size for the original deep learning algorithm. In the training process, the parameters of the deep learning algorithm were gradually adjusted mathematically to minimize the error between the actual and output values. Hold-out validation was used to verify the validity of the algorithm. The training dataset was used to train the CNN models, the validation dataset to select the most optimal CNN model during training, and the test dataset to evaluate the performance of the most optimal CNN model.
The training dataset contained more images of GISTs and leiomyomas than schwannomas, NETs, and ectopic pancreases. To overcome the imbalance in the data volume, the following methods were used. For the images of schwannomas, the images of the lesions were used as input values, and new images with the features of the original images were generated using a deep convolutional generative adversarial network (DCGAN) [34] as a preprocessing step in the training process of deep learning and used as the training data. For the images of ectopic pancreases, the images of the cases diagnosed on the basis of imaging findings (without pathological diagnoses) were used as unlabeled data, and relearning was performed using semi-supervised learning after training with labeled data.
To accelerate the training, super-computing resources (Flow, Information Technology Center, Nagoya University): Distributed-Data-Parallel (DDP) on multi-node, multi-process were used. To prevent overfitting, data augmentation, early stopping, and label smoothing were used. For data augmentation, image rotation (− 90, 0, 90), horizontal flip, vertical flip, random erasing, and RandAugment (N = 3, M = 4) were performed [35]. Stochastic gradient descent (SGD) with momentum was used to train the network weights as the optimization algorithm. A batch size of 288, 2000 epochs and a learning rate of 0.004 were used as the training parameters. The learning rate was decayed with cosine annealing, which is a type of learning rate schedule.
The trained AI system outputs a probability distribution (such that the sum of the scores for the five categories of SELs, i.e., GIST, leiomyoma, schwannoma, NET, and ectopic pancreas, would equal one) for each image. The predictive value of the differential diagnosis of SELs for each image was defined as the category of SELs with the highest probability score without a cutoff value. The predictive value for each case was defined as the mode value of the predictive values for all the images from each case.
Outcome measures
The main outcome measurements were the diagnostic performance of the AI system for the five category classification (GIST, leiomyoma, schwannoma, NET, and ectopic pancreas), differentiating GISTs from non-GISTs, and differentiating GIST/schwannoma from other SELs. The secondary outcome measurement was the diagnostic performance of the endoscopists. Among the four endoscopists, two experts were board-certified specialists at the Japan Gastroenterological Endoscopy Society. The experts had more than 10 years of experience in evaluating gastrointestinal SELs and had conducted more than 500 EUS examinations of gastrointestinal SELs. Two non-experts had 4–5 years of experience and had conducted less than 200 EUS examinations. The endoscopists were provided with all images from each case in the test dataset and asked to diagnose SELs as GIST, leiomyoma, schwannoma, NET, or ectopic pancreas for each case.
Statistical methods
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy were calculated to compare the performance of differentiating GISTs from non-GISTs and differentiating GIST/schwannoma from other SELs between the AI system and the endoscopists. The accuracies were calculated to compare the performance of the five-category classification between the AI system and the endoscopists. A two-sided McNemar test with a significance level of 0.05, was used to compare differences in accuracy, sensitivity, and specificity. Continuous variables were expressed as medians and ranges. Categorical variables were expressed as percentages. The Kruskal–Wallis test was used for continuous variables, and Fisher’s exact test was used for categorical variables. Statistical significance was set at P < 0.05; all tests were two-sided. Interobserver agreement of the endoscopists was assessed using kappa statistics: κ > 0.8, almost perfect agreement; 0.8–0.6, substantial agreement; 0.6–0.4, moderate agreement; 0.4–0.2, fair agreement; and < 0.2, slight agreement. A κ value of 0 indicated agreement equal to chance, and < 0 indicated disagreement. SPSS version 27.0 (IBM Japan Ltd, Tokyo, Japan) was used for all statistical analyses.
Results
Characteristics of the patients
Figure 1 shows a patient flowchart of the study. In this study, 243 SEL cases pathologically confirmed at the NUH, 392 SEL cases pathologically confirmed at 11 other participating hospitals, and 34 ectopic pancreas cases diagnosed on the basis of imaging findings (without pathological diagnoses) at the NUH were identified. After excluding cases according to the exclusion criteria (poor quality images; 3 SEL cases at the NUH, 1 SEL case at 11 other participating hospitals, and 1 ectopic pancreas case without a pathological diagnosis at the NUH), a total of 16,110 images were collected from 631 cases of pathologically confirmed SELs and 3179 images were collected from 33 cases of ectopic pancreases without pathological diagnoses. After selection, 11,305 images from 419 cases (6023 images from 287 GIST cases, 3771 images from 70 leiomyoma cases, 441 images from 14 schwannoma cases, 644 images from 34 NET cases, and 426 images from 14 ectopic pancreas cases) were used as the training dataset, 1930 images from 90 cases (1344 images from 63 GIST cases, 309 images from 13 leiomyoma cases, 192 images from 8 schwannoma cases, 54 images from 5 NET cases, and 31 images from 1 ectopic pancreas case) were used as the validation dataset, and 2875 images from 122 cases (1999 images from 85 GIST cases, 215 images from 14 leiomyoma cases, 274 images from 11 schwannoma cases, 198 images from 8 NET cases, and 189 images from 4 ectopic pancreas cases) were used as the test dataset. In the training dataset, 441 images from 14 schwannoma cases were used as input values, and new 40,000 images were generated using DCGAN. Then, 3000 images were manually extracted from these images and used as training data. The dataset of ectopic pancreases without pathological diagnoses was used as an unlabeled dataset for semi-supervised learning. Detailed clinical characteristics of the patients in the training, validation, test, and unlabeled ectopic pancreas datasets are shown in Table 1. There were no significant differences in age, sex, lesion location, lesion size on EUS images, and pathological type among the training, validation, and test datasets.
Flowchart of the study. A total of 631 cases of pathologically confirmed SELs were divided into development and test datasets, and 33 cases of ectopic pancreases without pathological diagnoses were used as an unlabeled dataset for semi-supervised learning. NUH Nagoya University Hospital, SELs subepithelial lesions, GIST gastrointestinal stromal tumor, NET neuroendocrine tumor, AI artificial intelligence
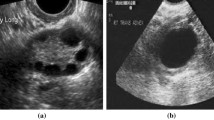
Examples of the test images are shown in Fig. 2. The AI system marked the SEL with a green square and showed the image-wise predictive value of the differential diagnosis of SELs, the probability score, and the case-wise predictive value.
Output of the artificial intelligence (AI) system. a An example of a gastrointestinal stromal tumor (GIST) case. b The AI system correctly diagnosed the lesion as GIST by indicating it with a green square frame. It showed the image-wise predictive value (GIST), the probability score (0.798), and the case-wise predictive value (mode GIST)
Five-category classification performance
The accuracies of the AI system and the expert with the best performance for the five-category classification (GIST, leiomyoma, schwannoma, NET, and ectopic pancreas) were 86.1% and 68.0%, respectively. The confusion matrix for the per-category diagnostic performance of the AI system and endoscopists is presented in Table 2. The accuracy of the AI system was significantly higher than that of all the endoscopists (accuracy range 27.0–68.0%, all the endoscopists P < 0.001). The accuracies of the experts were higher than those of the non-experts (54.9–68.0% vs. 27.0–46.7%). The per-category sensitivities of the AI system and experts were highest for GIST (98.8% and 63.5–77.6%, respectively) and lowest for schwannoma (45.5% and 0%, respectively). Both the AI system and experts identified schwannomas as GISTs in most misdiagnosed cases. The per-category sensitivity of the AI system for GIST was higher than that of all the endoscopists (98.8% vs. 25.9–77.6%).
The interobserver agreement among the experts for the five-category classification was fair (Fleiss’ κ = 0.382) and that among the non-experts was slight (Fleiss’ κ = 0.100).
Binary classification performance
The sensitivity, specificity, PPV, NPV, and accuracy of the AI system for differentiating GISTs from non-GISTs were 98.8%, 67.6%, 87.5%, 96.2%, and 89.3%, respectively. The detailed diagnostic performances of the AI system and endoscopists are shown in Table 3. The sensitivity and accuracy of the AI system were significantly higher than those of all the endoscopists (sensitivity: 98.8% vs. 25.9–77.6%, all the endoscopists, P < 0.001; accuracy: 89.3% vs. 44.3–71.3%, all the endoscopists, P < 0.001). The specificity of the AI system was comparable to or higher than those of the experts (67.6% vs 56.8–67.6%). Although the specificities of the non-experts (75.7–86.5%) were higher than those of the AI system and experts, the sensitivities and accuracies of the non-experts (25.9–54.1% and 44.3–60.7%, respectively) were lower than those of the AI and experts (63.5–77.6% and 64.8–71.3%, respectively).
The interobserver agreement among the experts for differentiating GISTs from non-GISTs was fair (Fleiss’ κ = 0.382) and that among the non-experts was slight (Fleiss’ κ = 0.045).
The sensitivity, specificity, PPV, NPV, and accuracy of the AI system for differentiating GIST/schwannoma from other SELs were 100.0%, 76.9%, 94.1%, 100.0%, and 95.1%, respectively. The detailed diagnostic performances of the AI system and endoscopists are shown in Table 4. The sensitivity and accuracy of the AI system were significantly higher than those of the expert with the best performance for differentiating GIST/schwannoma from other SELs (sensitivity: 100.0% vs. 84.4%, P < 0.001; accuracy: 95.1% vs. 82.8%, P = 0.003) and were significantly higher than those of all the endoscopists (sensitivity: 100.0% vs. 35.4–84.4%; accuracy: 95.1% vs. 42.6–82.8%). The specificity of the AI system was comparable to that of the experts (76.9%) and higher than that of the non-experts (50.0–69.2%). The specificities of the experts were higher than those of the non-experts. The sensitivity and accuracy of the expert with the best performance for differentiating GIST/schwannoma from other SELs were higher than those of the non-experts (sensitivity: 84.4% vs. 35.4–69.8%; accuracy: 82.8% vs. 42.6–67.2%), but the sensitivity and accuracy of the other expert were comparable to or lower than those of the non-expert with the best performance for differentiating GIST/schwannoma from other SELs (sensitivity: 64.6% vs. 69.8%; accuracy: 67.2% vs. 67.2%).
The interobserver agreement among the experts for differentiating GIST/schwannoma from other SELs was fair (Fleiss’ κ = 0.346) and that among the non-experts was disagreement (Fleiss’ κ = − 0.017).
Discussion
In this multicenter study, we developed an AI system with high diagnostic performance to classify upper gastrointestinal SELs on EUS images into multiple categories. This is the largest study regarding AI-based diagnostic systems of SELs from EUS images and the first study to apply AI for multi-category classifications of common types of SELs, such as GIST, leiomyoma, schwannoma, NET, and ectopic pancreas. This study included images obtained from EUS examinations performed using various types of echoendoscopes and ultrasound systems. This AI system may be useful in assisting endoscopists in predicting the pathological diagnosis of SELs during EUS examinations and in making better diagnostic and therapeutic decisions.
Regarding the efficacy of AI-based diagnostic systems of SELs on EUS images, Minoda et al. reported that the accuracies of the AI system for differentiating GISTs from non-GISTs were 86.3% in 30 images from 30 SELs < 20 mm and 90.0% in 30 images from 30 SELs ≥ 20 mm [29]. Yoon et al. reported that in the test datasets of 212 images from 69 SELs, the image-based accuracy of the AI system for differentiating GISTs from non-GISTs was 79.2%, and after further sequential analysis of 98 images classified as non-GISTs with the AI system to differentiate leiomyomas from schwannomas, the final accuracy was 75.5% [30]. However, because of the limited number of cases included in previous studies and the application of AI for binary classification only, these values should be carefully considered, and their utility in the real clinical practice seems to be limited. The accuracy of our AI system for differentiating GISTs from non-GISTs (89.3%) was comparable to or higher than that of previous studies. We have developed an AI system for multi-category classifications of common types of SELs for a more clinically relevant differential diagnosis, and an accuracy of 86.0% was considered acceptable for use in clinical practice.
In this study, we compared the diagnostic performance of the AI system with that of experts and non-experts. The accuracy of the endoscopists for the five-category classification in this study ranged from 27.0 to 68.0%, which was comparable to the reported accuracy of the differential diagnosis of SELs (45.5–66.7%) [11,12,13]. The accuracy of the endoscopists for differentiating GISTs from non-GISTs in this study ranged from 44.3 to 71.3%, which was also comparable to that in previous studies (53.3–75.9%) [29, 30]. Overall, the diagnostic performance of the AI system was higher than that of all the endoscopists, and that of the experts was higher than that of the non-experts. The interobserver agreement in this study was not high because of the subjective interpretation of EUS imaging findings, and the length of experience with EUS was important for the accuracy of SELs diagnosis. These results are similar to those of the previous studies [36], and the AI system may be especially useful for non-experts to predict the pathological diagnosis of SELs during EUS examinations. In the five-category classification, both the AI system and experts identified schwannomas as GISTs in most misdiagnosed cases. The differentiation of schwannomas from GISTs with different prognoses was difficult even for experts, and this is consistent with previous studies [9, 10]. The AI system, with comparable specificity and higher sensitivity and accuracy than those of the experts for differentiating GISTs from non-GISTs and differentiating GIST/schwannoma from other SELs, can be useful in determining the need for tissue sampling or surgical resection after EUS examinations.
This study is unique in that it applies DCGAN and semi-supervised learning to training images of schwannomas and ectopic pancreases. Because these lesions are rare, and biopsy is not recommended for undoubtedly benign SELs such as leiomyomas and ectopic pancreases diagnosed based on computed tomography (CT) and/or EUS findings [3, 4, 16], the number of pathologically confirmed lesions is limited. In addition to training with pathologically confirmed SEL images, training with new images with features of the original schwannoma images generated using DCGAN and relearning with images of ectopic pancreas cases diagnosed on the basis of imaging findings (without pathological diagnoses) as unlabeled data using semi-supervised learning were thought to contribute to improving the diagnostic performance of the AI system.
This study included five common types of SELs and did not include other rare types of SELs. Therefore, the prevalence of GISTs in this study (68.5%) was higher than that in previous studies on the diagnosis of SELs by EUS, EUS-FNAB, and mucosal cutting biopsy (39.2–67.0%) [12, 17,18,19,20,21]. This may have affected the PPVs and NPVs of the AI system and endoscopists for differentiating GISTs from non-GISTs and differentiating GIST/schwannoma from other SELs. We are planning to include other types of SELs to confirm our results.
Due to the limited number of non-GIST cases, this study focused on GISTs and schwannomas in the binary classification performance based on prognosis and treatment strategy. Before clinical application, we are planning to investigate the diagnostic performance in classifications that would be more useful for making therapeutic decisions, such as differentiating GIST/NET from other SELs and differentiating GISTs from schwannomas.
This study has several limitations. First, there might be discrepancies between the pathological diagnosis of biopsy specimens and actual pathological diagnosis in SELs that were not surgically or endoscopically resected, and between the image diagnosis and actual pathological diagnosis in the ectopic pancreas cases used as unlabeled data. However, in these cases, immunohistochemical analysis results and/or periodic follow-up examinations may have reduced the discrepancies. Second, all EUS images including test datasets were still images obtained retrospectively from affiliated hospitals without randomization, making it difficult to exclude selection bias. Therefore, the diagnostic performance of the AI system might be overestimated. In this study, training, validation, and test datasets were divided using random sampling. However, the generalizability of the system should be evaluated with real-time external validation. Some EUS images obtained from older endoscopy systems had lower resolution than recently adopted systems, which may have affected the diagnostic performance of the AI system and endoscopists. Third, although we applied DCGAN and semi-supervised learning to overcome the imbalance between the training data volume in GISTs and non-GISTs, the number of non-GIST cases included in the test dataset may have been insufficient to evaluate the diagnostic performance of the system. Large-scale multicenter prospective studies are needed to develop graphical user interfaces that can be used in real clinical practice and to confirm the diagnostic performance of the system, using not only still EUS images but videos obtained from non-affiliated hospitals as well.
In conclusion, the AI system for classifying upper gastrointestinal SELs showed higher diagnostic performance than that of the experts. It may assist endoscopists in improving the diagnosis of SELs in clinical practice.
References
Hedenbro JL, Ekelund M, Wetterberg P. Endoscopic diagnosis of submucosal gastric lesions. The results after routine endoscopy. Surg Endosc. 1991;5:20–3.
Park CH, Kim EH, Jung DH, Chung H, Park JC, Shin SK, Lee YC, et al. Impact of periodic endoscopy on incidentally diagnosed gastric gastrointestinal stromal tumors: findings in surgically resected and confirmed lesions. Ann Surg Oncol. 2015;22:2933–9.
Faulx AL, Kothari S, Acosta RD, Agrawal D, Bruining DH, Chandrasekhara V, et al. The role of endoscopy in subepithelial lesions of the GI tract. Gastrointest Endosc. 2017;85:1117–32.
Kim GH, Park do Y, Kim S, Kim DH, Choi CW, Heo J, et al. Is it possible to differentiate gastric GISTs from gastric leiomyomas by EUS? World J Gastroenterol. 2009;15:3376–81.
Rubin BP, Heinrich MC, Corless CL. Gastrointestinal stromal tumour. Lancet. 2007;369:1731–41.
Nishida T, Hirota S. Biological and clinical review of stromal tumors in the gastrointestinal tract. Histol Histopathol. 2000;15:1293–301.
Daimaru Y, Kido H, Hashimoto H, Enjoji M. Benign schwannoma of the gastrointestinal tract: a clinicopathologic and immunohistochemical study. Hum Pathol. 1988;19:257–64.
Voltaggio L, Murray R, Lasota J, Miettinen M. Gastric schwannoma: a clinicopathologic study of 51 cases and critical review of the literature. Hum Pathol. 2012;43:650–9.
Mekras A, Krenn V, Perrakis A, Croner RS, Kalles V, Atamer C, et al. Gastrointestinal schwannomas: a rare but important differential diagnosis of mesenchymal tumors of gastrointestinal tract. BMC Surg. 2018;18:47.
Lauricella S, Valeri S, Mascianà G, Gallo IF, Mazzotta E, Pagnoni C, et al. What about gastric schwannoma? A review article. J Gastrointest Cancer. 2021;52:57–67.
Karaca C, Turner BG, Cizginer S, Forcione D, Brugge W. Accuracy of EUS in the evaluation of small gastric subepithelial lesions. Gastrointest Endosc. 2010;71:722–7.
Lim TW, Choi CW, Kang DH, Kim HW, Park SB, Kim SJ. Endoscopic ultrasound without tissue acquisition has poor accuracy for diagnosing gastric subepithelial tumors. Medicine. 2016;95:e5246.
Hwang JH, Saunders MD, Rulyak SJ, Shaw S, Nietsch H, Kimmey MB. A prospective study comparing endoscopy and EUS in the evaluation of GI subepithelial masses. Gastrointest Endosc. 2005;62:202–8.
Akahoshi K, Oya M, Koga T, Shiratsuchi Y. Current clinical management of gastrointestinal stromal tumor. World J Gastroenterol. 2018;24:2806–17.
Casali PG, Abecassis N, Aro HT, Bauer S, Biagini R, Bielack S, et al. Gastrointestinal stromal tumours: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:68–78.
Nishida T, Blay JY, Hirota S, Kitagawa Y, Kang YK. The standard diagnosis, treatment, and follow-up of gastrointestinal stromal tumors based on guidelines. Gastric Cancer. 2016;19:3–14.
de Moura DTH, McCarty TR, Jirapinyo P, Ribeiro IB, Flumignan VK, Najdawai F, et al. EUS-guided fine-needle biopsy versus fine-needle aspiration in the diagnosis of subepithelial lesions: a large multicenter study. Gastrointest Endosc. 2020;92:108–19.
Ihara E, Matsuzaka H, Honda K, Hata Y, Sumida Y, Akiho H, et al. Mucosal-incision assisted biopsy for suspected gastric gastrointestinal stromal tumors. World J Gastrointest Endosc. 2013;5:191–6.
Minoda Y, Chinen T, Osoegawa T, Itaba S, Kazuhiro H, Akiho H, et al. Superiority of mucosal incision-assisted biopsy over ultrasound-guided fine needle aspiration biopsy in diagnosing small gastric subepithelial lesions: a propensity score matching analysis. BMC Gastroenterol. 2020;20:19.
Akahoshi K, Oya M, Koga T, Koga H, Motomura Y, Kubokawa M, et al. Clinical usefulness of endoscopic ultrasound-guided fine needle aspiration for gastric subepithelial lesions smaller than 2 cm. J Gastrointest Liver Dis. 2014;23:405–12.
Larghi A, Fuccio L, Chiarello G, Attili F, Vanella G, Paliani GB, et al. Fine-needle tissue acquisition from subepithelial lesions using a forward-viewing linear echoendoscope. Endoscopy. 2014;46:39–45.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Fujioka T, Kubota K, Mori M, Kikuchi Y, Katsuta L, Kasahara M, et al. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Jpn J Radiol. 2019;37:466–72.
Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–23.
Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25–32.
Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653–60.
Byrne MF, Chapados N, Soudan F, Oertel C, Linares Perez M, Kelly R, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100.
Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, et al. Usefulness of deep learning analysis for the diagnosis of malignancy in intraductal papillary mucinous neoplasms of the pancreas. Clin Transl Gastroenterol. 2019;10:1–8.
Minoda Y, Ihara E, Komori K, Ogino H, Otsuka Y, Chinen T, et al. Efficacy of endoscopic ultrasound with artificial intelligence for the diagnosis of gastrointestinal stromal tumors. J Gastroenterol. 2020;55:1119–26.
Kim YH, Kim GH, Kim KB, Lee MW, Lee BE, Baek DH, et al. Application of a convolutional neural network in the diagnosis of gastric mesenchymal tumors on endoscopic ultrasonography images. J Clin Med. 2020;9:3162.
Lee DH. Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks. ICML Work Chall Represent Learn. 2013;3(2):1–6.
Tan M, Le QV (2021) EffIicientNetV2: Smaller models and faster training. In: Proceedings of the 34th international conference on machine learning, PMLR 139:10096–10106
Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst. 2012;25:1097–105.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27:2672–80.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6:60.
Gress F, Schmitt C, Savides T, Faigel DO, Catalano M, Wassef W, et al. Interobserver agreement for EUS in the evaluation and diagnosis of submucosal masses. Gastrointest Endosc. 2001;53:71–6.
Acknowledgements
We thank Dr. Tatsuya Kawamura and Dr. Hidetaka Takahashi in Nagoya University Graduate School of Medicine for their help with this study. This work was supported in part by a grant from the Shimadzu Foundation for the Advancement of Science and Technology. The super-computing resources were produced by Information Technology Center, Nagoya University.
Funding
This study was funded by the Shimadzu Foundation for the Advancement of Science and Technology.
Author information
Authors and Affiliations
Contributions
KH designed the study, acquired and analyzed the data, and drafted the manuscript. TK analyzed the data and assisted with drafting the manuscript. KF, NK, SF, HY, TM, HA, KM, YS, DS, KY, TN, DH, TO, TK, EI, TS, KM, TY, TI, EO, MN, HK, and MI were involved in the acquisition of the data. KF and MF revised the manuscript. MF gave the final approval of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Kazuhiro Furukawa has received a research grant from the Shimadzu Foundation for the Advancement of Science and Technology. Mitsuhiro Fujishiro received research grants from Fujifilm Corporation, HOYA Corporation, and Olympus Corporation, and honoraria from Fujifilm Corporation and Olympus Corporation outside this study. All other authors declare that they have no conflict of interest.
Human rights statement
All procedures followed were in accordance with the Ethical Standards of the Responsible Committee on Human Experimentation (Institutional and National) and with the Declaration of Helsinki of 1964 and later versions.
Informed consent
The written consent was waived by the ethics committees because the study had a retrospective design.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MP4 17254 KB)
Rights and permissions
About this article
Cite this article
Hirai, K., Kuwahara, T., Furukawa, K. et al. Artificial intelligence-based diagnosis of upper gastrointestinal subepithelial lesions on endoscopic ultrasonography images. Gastric Cancer 25, 382–391 (2022). https://doi.org/10.1007/s10120-021-01261-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-021-01261-x