Abstract
Crohn’s disease and ulcerative colitis are two chronic diseases that cause inflammation in the tissues of the entire gastrointestinal tract and are described by the term inflammatory bowel disease. Gastroenterologists find it difficult to evaluate endoscopic images to recognise the characteristics of the two chronic diseases. Therefore, this work aims to build a dataset with images of Crohn’s disease and ulcerative colitis (collected from the public datasets LIMUC, HyperKvasir and CrohnIPI) and train deep learning models (five CNNs and six ViTs) to develop a tool capable of helping doctors to distinguish the type of inflammatory bowel disease. In addition, as these architectures will be too heavy to work in a hospital context, in this work, we are looking to use knowledge distillation to create lighter and simpler architectures with the same precision as the pre-trained architectures used in this study. During this process, it is important to evaluate and interpret the pre-trained architectures before the distillation process, and the architectures resulting from knowledge distillation to ensure that we can maintain performance and that the information learnt by both architectures are similar. It is concluded that is possible to reduce 25x the number of parameters while maintaining good performance and reducing the inference time by 5.32 s. Allied with this, through the interpretability of the models was concluded that both before and after the knowledge distillation are possible to identify ulcers, bleeding situations, and lesions caused by the inflammation of the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Inflammatory bowel disease (IBD) is the term used to classify two chronic diseases: ulcerative colitis and Crohn’s disease, which cause digestive derangements and inflammation in the entire gastrointestinal tract, from the mouth to the anus. So far, its cause is scientifically unknown. However, some environmental factors that interact with genetics cause a reaction in the immune system [1, 2]. Crohn’s disease is a chronic inflammatory disease that causes inflammation in the mucosa of the intestine, most commonly in the lower part of the small intestine, but can cause inflammation anywhere in the gastrointestinal tract, from the mouth to the anus. It occurs in patients aged 15–35 years and causes pain, diarrhoea, fever, and other symptoms. On the other hand, ulcerative colitis is associated with blood in the stool, intense pain, and diarrhoea. Unlike Crohn’s disease (CD), ulcerative colitis (UC) affects the mucosal layer of the colon, causing lesions in the large intestine and rectum [3].
The number of patients with this condition (IBD) is increasing worldwide and can affect people of all ages, including children and the elderly. Unlike other inflammatory diseases, there is no medical treatment that can cure inflammation in the intestine. However, an accurate and early diagnosis allows the gastroenterologist to prescribe a treatment that minimises the symptoms of the disease and offers a quality of life to patients. In addition, if the inflammation is not treated in time, it can lead to colon cancer and damage to the walls of the intestine [4, 5].
The use of deep learning models to help the gastroenterologist in the diagnosis of patients with inflammatory bowel disease is important and, with time, more necessary. Not only in Portugal but worldwide, the number of people who have this chronic disease is increasing, and its diagnosis depends on the evaluation that the gastroenterologist makes of the endoscopic images, which makes this diagnosis subject to a great subjectivity [3]. Therefore, it is crucial to find tools that automate the diagnosis of the disease to prescribe the most suitable treatment for the patient.
The literature has focused on the implementation of deep learning models in image processing of Crohn’s disease or ulcerative colitis disease. Where, for example, the authors [6] sought to diagnose endoscopic mucosal healing in ulcerative colitis using deep learning and machine learning algorithms. In this study, 856 endoscopic images with MES 0–3 were used, and two experiments were performed: (i) classify MES 0–1 vs MES 2–3 images to detect the presence or absence of mucosal healing and (ii) classify MES 0 vs MES 1 images to determine complete or incomplete healing. Also, the literature in [7] used the HyperKvasir dataset to obtain 840 images of ulcerative colitis disease, to diagnose the existence of the disease by classifying it with the others in the gastrointestinal tract, and to identify the degree of severity of ulcerative colitis. Another study developed by [8] consisted of using a CNN to identify and differentiate multiple lesions in the small intestine with distinct bleeding potentials, without specifying which disease they sought to classify.
Most of the studies developed by other authors focus on a single inflammatory bowel disease (either ulcerative colitis or Crohn’s disease) and are very useful tools to help doctors to monitor the progression of the disease. However, this work aims to develop a solution that helps doctors in the initial phase, which is the diagnosis of inflammatory bowel disease. Allied to this, we also consider it important to use the interpretability of the models, because in this way, the tool can provide information about inflammatory bowel disease, as well as showing which parts of the intestine have been considered for prediction.
This work makes a scientific contribution to recognising which type of inflammatory bowel disease is present: Crohn’s disease or ulcerative colitis, using colonoscopy and video capsule endoscopy to gather images, using six convolutional neural networks (CNNs) and five vision transformers (ViTs) to understand which of the deep learning architectures is best. The computational demands of these deep learning models that have been pre-trained on large volumes of data were also taken into consideration. The training of architectures with 25x fewer parameters was undertaken by distilling the knowledge of the pre-trained architectures. In this way, it is possible to have lighter architectures with the same precision as the main architectures [9]. Finally, it is relevant to the study to evaluate the models qualitatively and quantitatively before and after the knowledge distillation process to understand what information was learnt by both architectures (qualitative evaluation) and whether the performance of the models remained the same after the process (quantitative evaluation). This work is an extension of the paper published at IbPRIA 2023 [10]. The differences between the papers stand out by:
-
1.
Creation of one more dataset;
-
2.
Three more experiences were realised;
-
3.
Implementation of two techniques of knowledge distillation;
-
4.
Implementation of vision transformers for classification of the images;
-
5.
Interpretability of the models before and after knowledge distillation;
-
6.
The ensemble model (ResNet50+MobileNetV2) was modified.
In summary, the focus of the paper [10] was to build a tool using deep learning models to help doctors diagnose IBD. The contributions of this paper were the development of six experiments and the publication of the quantitative results of six pre-trained CNNs. Later, in the paper [11] and acknowledging the increasing popularity of ViTs [12], the same methodology was used, but with the addition of five ViTs so that we could compare the quantitative results of both architectures. Also, the distillation of knowledge from the pre-trained architectures of CNNs and ViTs was applied. For the present paper, three more experiments were developed. We chose the best experiment from those published in previous papers and those developed in this study, in order to publish the best quantitative results before and after the knowledge distillation process. Finally, the qualitative results (interpretability) of the best model are included to demonstrate its ability to recognise the characteristics associated with the two types of IBD.
The organisation of this document is divided into five sections: Section 2 describes the findings used in the literature review; Section 3 describes the created dataset for this study and the implemented experimental methodology; Section 4 presents the quantitative results obtained by the teachers and student’s models; Section 5 presents the information obtained after the interpretability of the models; and Section 6 summarises the findings, suggests some future research directions, and presents the strengths and limitations of the work.
2 Literature review
Over time, the integration of computer vision tools in the diagnosis of inflammatory bowel disease has grown increasingly crucial. Given that the assessment of images acquired through medical imaging procedures relies on the judgement of expert gastroenterologists, the criteria for determining a patient’s diagnosis remain subjective and may vary among healthcare professionals [13]. This situation, in turn, has implications for patients, leading to delays in receiving timely and effective treatment for their condition.
2.1 Research methodology
2.1.1 Data sources
PubMed and Google Scholar were chosen as the data sources to extract the primary studies. The number of results found after searching papers in each of the data sources is shown in Table 1.
2.1.2 Search string
Based on the aim of this study, some search strings, tunned for each selected data source, were developed. The search was also made by title. Table 2 provides a list of the search strings and titles used in each electronic database.
2.1.3 Inclusion criteria
The inclusion criteria set to select the articles were that studies had to be published between January 2016 and December 2022. In addition to this, the studies had to address the use of deep learning to distinguish inflammatory bowel diseases. Whenever this was not possible, at least, they had to apply deep learning to classify endoscopic or colonoscopy images of the disease.
2.1.4 Exclusion criteria
Studies that directed their research towards using deep learning to classify other bowel diseases were excluded as polyps, haemorrhoids, etc. and also were excluded articles that were duplicated with those that had already been collected. Papers that oriented the investigation to processing videos collected during colonoscopy or endoscopy were also discarded.
2.1.5 Results
Accounting for all the papers selected on each of the searches performed, 48 papers were collected. In the first approach, seven papers were discarded because they were repeated, and seven more papers were discarded because they had few citations. Then, by reading the abstract, seven more papers were discarded because they were not within the scope of the study of this work. In total, 11 papers were counted for the literature review. Figure 1 shows the distribution of papers by year of publication. Table 3 lists all the papers that were selected for this literature review.
2.2 Findings
Table 4 presents an overview of the results obtained and the architectures used by the literature in the field of inflammatory bowel disease diagnosis. As a result, this section will provide a summary of the information included in the gathered papers.
The authors in [17] conducted their study to use deep learning algorithms to distinguish between active and non-active inflammation of Crohn’s disease in patients. For the study, the authors collected 6,205 images from 54 patients, which are divided into 32 patients with active Crohn’s disease and 22 patients who form a control group. However, the authors in the control group enclose images of patients who do not have any inflammation and patients who have had inflammation. In this study, a CNN+LSTM was combined, and a CNN was used to make the automatic diagnosis. The combination of a CNN+LSTM is the best model to do this with 95.36% of accuracy (Acc), 94.6% of sensitivity, and 92.78% of specificity. However, it is not shown which CNN was used.
In the study developed in [21], the authors sought to classify whether ulcerative colitis disease was in remission or not. They also wanted to automatise the classification of the severity grade of UC based on Mayo scores 1–3. Mayo 0 images were considered healthy patients, which is not true because patients with Mayo 0 have already been diagnosed with ulcerative colitis disease, but at that moment, the disease is in remission/inactive. The authors in this study used colonoscopy images collected from the First Affiliated Hospital of Kunming Medical University. They propose an architecture that combines a CNN and an RNN, called UC-DenseNet, and compare the results with other architectures: VGG-19, ResNet152, InceptionV3, and DenseNet201. The proposed architecture stands out obtaining 97.6% of Acc, 97.5% of Pre, 98.0% of recall, 97.6% of F1-Score, and 97.5% of AUC for the classification of the existence of remission or not. An Acc=90.6%, Pre=87.4%, recall=86.3%, and F1-Score=86.8% were obtained for the grade of disease severity.
The literature in [20] collected 57,597 colonoscopy images about Crohn’s disease, ulcerative colitis, and from healthy patients. However, this dataset is not publicly available. The authors aimed to use 13,872 images to train the CNN ResNetXt-101 network and hold 1,452 images for testing. In which, they aimed to compare with the analysis done by six experienced clinicians. The comparison consisted of analysis per image of the classes, but also per patient to evaluate the diagnosis made. Based on the results obtained, the CNN for the classification of CD images obtained 92.39% of Acc, 87.53% of sensitivity, 94.78% of specificity, 89.19% of PPV, and 88% of F1-Score. For UC, it obtained: 93.35% of Acc, 90.49% of sensitivity, 98.14% of specificity, 89.94% of PPV, 95.11% of NPV, and 90% of F1-Score. They concluded that the CNN was better than clinicians at classifying images. On the other hand, clinicians were able to be better most of the time in per-patient analysis.
The study in [15] aimed to detect Crohn’s disease lesions caused by the different degrees of severity located in the small intestine and colon. The authors collected 7,744 images from 38 patients, from three centres in South-east Denmark. They used the ResNet50 network in five different image pre-processing (e.g. Original, Contrast increase, Histogram equalisation, GradientX, and Dephezing); this pre-processing consisted of modifying the texture of the images to improve the performance of the network. To evaluate the performance of the network, the authors divided the dataset by patient and random which consisted of 70% for training, 10% for validation, and 20% for testing. They concluded that overall for the per-patient split, the CNN obtained 98.30% of accuracy, 95.72% of sensitivity, and 99.77% of specificity. In the random split, it obtained 98.58% of Acc, 96.21% of sensitivity, and 100% of specificity.
Stidham et al. [13] aimed to compare the performance of a deep learning model with 10 human reviewers for the classification of the severity grade of ulcerative colitis disease. For the study, 16,514 images were used as the Mayo score 0–3. The dataset was divided into 90% for creating the model and 10% for testing; of the 90%, 80% was used for training and 10% for fine-tuning. The authors demonstrated that the InceptionV3 network performed very similarly to the reviewers when comparing the values of the Kappa statistics (k) metric, where the network obtained k=0.84 and the experienced reviewers obtained k=0.86. Also, they concluded that the model demonstrated a good ability to distinguish endoscopic remission (Mayo 0–1) from moderate-to-severe (Mayo 2–3) with an AUROC of 0.966, a PPV of 0.87, a sensitivity of 83.0%, a specificity of 96.0%, and an NPV of 0.94.
The authors in [16] aimed to automate the detection of intestinal strictures in Crohn’s disease. In this study, the authors also used images of normal mucosal and mucosal ulcers. In total, the dataset contained 27,892 images gathered by capsule endoscopy. To evaluate the performance of the network, the authors split the dataset using the 10-fold cross-validation method with seven patients for training and three for testing. To do the classification and imaging, the authors selected the EfficientNet-B5 network. It is shown in this study that the network can obtain an average accuracy of 93.5% for the classification of images of strictures and non-strictures. The authors further concluded that the network has an excellent differentiation between strictures and normal mucosa (AUC=0.989), strictures and all ulcers (AUC=0.942), and between strictures and different grades of ulcers (mild, moderate, and severe ulcers) with an AUC of 0.992, 0.975, and 0.829, respectively.
The study in [14] sought to gather images by a video capsule endoscopy to detect ulcers in Crohn’s disease. Therefore, the authors collected a dataset with 17,640 images from 49 patients and performed two experiments: (i) They divided the dataset into five subsets of equal size where 80% of the patients were for training and 20% for testing and (ii) they divided the dataset into N−1 patients for training and one unseen patient for testing, this process was randomised during 10 times. To classify the images, they used a CNN Xception network. In summary, the CNN in the first experiment obtained on average AUC=0.991, Acc=0.9612, sensitivity=0.9498, specificity=0.9698, PPV=0.958, and NPV=0.9642. In the second experiment, it averaged AUC=0.974, Acc=0.9066, sensitivity=0.8882, specificity=0.9077, PPV=0.9151, and NPV=0.9003.
The study developed in Mascarenhas et al. [8] consisted in using a CNN to identify and differentiate multiple lesions in the small intestine with distinct bleeding potentials. The authors used a dataset of 53,555 images, which they divided into 80% for training and 20% for validation to train the Xception network. Based on Saurin’s classification, the images were divided into three categories: no bleeding potential – P0; uncertain/intermediate bleeding potential – P1; and high bleeding potential – P2. In the end, the authors concluded that the CNN was able to differentiate multiple small bowel abnormalities and their respective bleeding potential classification with 99% of Acc, 88% of sensitivity, 99% of specificity, 87% of PPV, and 99% of NPV.
The study in [19] sought to collect coloscopy images from five hospitals in China to identify ulcerative colitis, Crohn’s disease, or healthy bowel. The authors were able to gather 49,154 colonoscopy images and also during the condition compared the performance of the model with 10 endoscopists. These images were only gathered for this study and are not publicly available. The team of human reviewers consisted of five reviewers with little experience and five very experienced reviewers. For the classification of the images, the authors used the ResNet50 network. When comparing the results of the deep learning model with the 10 reviewers, the authors showed that the model was on average faster at reading at 6.00 s and the reviewers with 2,425.00 s. On a per-patient analysis, the model was better than the reviewers with 0.991 of Acc versus 0.922 for the highly experienced reviewers and 0.780 for the less experienced reviewers. And in a per-lesion analysis, it was also superior with 0.904 of Acc against 0.597 for the less experienced reviewers and 0.699 for the very experienced reviewers.
The literature in [7] used the HyperKvasir dataset to obtain 840 images of ulcerative colitis disease, to diagnose the existence of the disease by classifying it with the others in the gastrointestinal tract, and to identify the degree of severity of ulcerative colitis. In this study, the authors used four convolutional networks: ResNet50, VGG-19, InceptionV3, and DenseNet121. Based on the experiments performed, the authors concluded that the four networks showed better performance in diagnosing ulcerative colitis and non-ulcerative colitis, where the VGG-19 network obtained 98.49% of Acc, 98.61% of sensitivity, 9824% of specificity, 97.66% of F1-Score, and 99.88% of AUC. On the other hand, the networks failed to achieve a good performance in grade severity classification, where DenseNet121 obtained 87.50% of Acc, 79.00% of sensitivity, 91.00% of specificity, 91.29% of F1-Score, and 90% of AUC.
The authors in the study [18] set a goal for their study to detect Crohn’s disease from ulcerative colitis on endoscopic images. Throughout the study, three classification tasks were performed: (i) distinguish the existence and non-existence of IBD; (ii) distinguish Crohn’s disease from ulcerative colitis; and (iii) distinguish ulcerative colitis from no ulcerative colitis. The authors used the SI-cura dataset, where they collected 4,388 images of ulcerative colitis, 5,948 images of Crohn’s disease, 1,067 images of IBD, and 2,822 of no pathology. The SI-cura project is an Italian initiative that seeks to develop solutions based on artificial intelligence to distinguish pathologies with different natures. This dataset was used for this project and was not made available. The networks ResNet18, ResNet34, ResNet50, ResNet101, and ResNet152 were chosen to classify the images. They also created an ensemble method that combines ResNet34, with ResNet50 and ResNet101. They concluded that the ensemble method was the best in diagnosing the existence or non-existence of IBD pathology with 0.940 of MCC, 1.00 of TNR, 0.968 of TPR, 0.912 of NPV, and 1.00 of PPV.
While traditional pipelines included a formal step of feature extraction [22], based on the recent findings made by other authors on the findings made by other authors, we seek to stand out by proposing to pool the images by the type of inflammatory bowel disease to distinguish between Crohn’s disease and ulcerative colitis, comparing ViTs with CNNs for processing the collected endoscopic images. A methodology that includes image pre-processing techniques for removing misleading features from endoscopic examination images is also suggested. Moreover, the present study also includes interpretability techniques to understand what regions of the mucosal the classifier identifies as the presence of the disease. Combined with this, we seek through knowledge distillation to build more lightweight architectures, possible to deploy in endoscopic systems.
3 Methodology
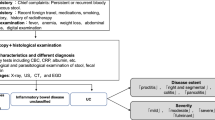
To enhance the diagnosis of inflammatory bowel disease, a methodology consisting of six phases is introduced, as depicted in Fig. 2. In the initial phase, images representing two types of inflammatory bowel diseases, namely ulcerative colitis and Crohn’s disease, were collected. This includes the extraction of frames from two videos recorded during endoscopic examinations conducted on patients, found in the HyperKavasir dataset. Also, a Gaussian Blur was applied to images and frames.
In the second phase, data augmentation was applied to the training set. Moving to the third phase, convolutional networks and vision transformers were implemented and configured. The performance assessment of these CNNs and ViTs models was conducted in the fourth phase, using various classification metrics such as accuracy, precision, recall, F1-Score, area under curve, and inference time. The fifth phase focused on attempting knowledge distillation for the deep learning models; once completed, they returned to the fourth phase to assess the performance of the models after the distillation process. Finally, the interpretability of both the models before and after the knowledge distillation process is performed.
This methodology is visually represented in Fig. 2 and will be elaborated upon in the subsequent sections.
3.1 Experimental setup
In conducting this study, Tensorflow, version 2.8.0, Tfimm, version 0.6.13, and Shap, version 0.40.0, were used. The programming environment for importing the libraries was Google Colab with the NVIDIA A100 GPU.
Three databases of images related to Crohn’s disease and Ulcerative colitis were used to conduct this study. With the aim of combining the images from the two diseases into a single dataset, the images referring to the Ulcerative Colitis pathology of the HyperKvasirr [23, 24] and LIMUC [25] databases will be used, as well as the images with the type of Crohn’s disease lesions present in the CrohnIPI database [26,27,28,29]. Figure 3 shows an example of the images that exist in each collected database.
Furthermore, the HyperKvasir database also comprises videos that have been reviewed and identified by gastroenterologists. The aim of including these videos is to acquire additional images associated with ulcerative colitis disease, thereby achieving a balanced representation of the classes within the dataset. To accomplish this, 20 frames per second were extracted from the videos, ensuring that these frames shared the same dimensions as the existing dataset images (\(572\times 531\)). This process yielded a total of 64 frames. One dataset was then created with:
-
446 instances of the LIMUC database, the instances are equally distributed by severity degree. That is, 25% of the total images referring to the severity degree were extracted from the original database; 64 video frames were extracted from the labelled videos from the HyperKvasir database; 850 instances from the HyperKavsir database; and 1360 instances from the CrohnIPI database. Figure 4 shows the class distribution of this dataset (Fig. 5).
In the images and frames extracted from the videos of the HyperKvasir database, a green square was observed in the lower left corner, indicating the endoscope’s position during diagnostic examinations. This presented a potential challenge when classifying the type of inflammatory bowel disease depicted in the images. To address this issue, a Gaussian blur with a radius of 2/16 was applied exclusively to the area containing the green square. An illustration of this transformation is provided in Figure 5.
3.2 Data augmentation
Prior to classifying the images, data augmentation was applied to the training set. This involved performing random flips in both horizontal and vertical directions, adjusting contrast randomly with a factor of 0.15, introducing random rotations with a factor of 0.2, and applying random zoom with a height portion of −0.2 and a width portion of −0.3.
3.3 Deep learning models
Based on the selected works during the literature review and in the advantages presented by [30], six CNNs were selected: AlexNet, VGG-16, ResNet50, InceptionV3, ResNet50+MobileNetV2, and a Hybrid model. Five ViTs were also selected: ViT-B/32, ViT-S/32, ViT-B/16, ViT-S/16, and the R26+S/32.
The architectures were trained for 200 epochs, using SparseCategoricalCrossentropy as the loss function, a batch size of 32, and Adam with a learning rate of 1.00e-05 was set as the optimizer [31]. During training, an EarlyStopping callback was used with the patience of 5 to monitor the validation accuracy [32, 33]. Default values were used for the remaining parameters, and no hyperparameter tuning was performed. The images were resized to 224×224 pixels.
3.3.1 Convolutional neural networks
The following architectures were used to classify images of inflammatory bowel diseases: AlexNet, ResNet50, VGG-16, and InceptionV3. These architectures have been pre-trained with the ImageNet dataset, except the AlexNet network because it was not possible to find the architecture pre-trained in Keras. Besides these architectures, another architecture was built that combines a ResNet50 with a MobileNetV2 network. Finally, a hybrid model was built where a CNN is combined with an LSTM. These architectures were chosen based on results presented in some studies of the literature review [7, 13, 15, 17, 18].
The fusion of the ResNet50 network and the MobileNetV2 network consists in an ensemble method, where the two networks were combined. The input images are directed to the two architectures that will process these images without the last classification layer, and the output of the two is concatenated to be classified by a Dense layer with two neurons referring to the classes Crohn’s disease and ulcerative colitis. The construction of this model was inspired by [34, 35]. This architecture is illustrated in Fig. 6.
The hybrid model built in this work was based on a similar architecture developed by other authors [17, 36, 37]. This architecture includes: eight Conv2D layers, two BatchNormalization layers, four MaxPooling2D layers, one Flatten layer, seven Dense layers, three Dropout layers, and three LSTM layers. The hybrid model is illustrated in Fig. 7. The forget gate in LSTM networks determines which information should be remembered and which should be forgotten before sending it to the subsequent layers [17].
3.3.2 Vision transformers
In addition to convolutional neural networks, transformer-based deep learning architectures were also used in this work, so that the two types of deep learning architectures could be compared in the task of classifying the type of inflammatory bowel disease. Therefore, in this section, the used vision transformers are presented [30].
Among the vision transformers, the following models were selected: ViT-B/32, ViT-S/32, ViT-B/16, ViT-S/16, and R26+S/32, as introduced in the works of [38,39,40]. These models were initially pre-trained using the Imagenet21k dataset. The R26+S/32 model, developed by Google and shared with the community, is one of the variants closely related to this approach. It involves a fusion of a ResNet network with a vision transformer, a concept described by [41]. In the model’s name, the “R26” portion signifies the number of convolutional layers incorporated from the ResNet network. The latter part, “S/32”, indicates the utilisation of these ResNet layers in combination with a compact vision transformer composed of 32 patches.
3.4 Knowledge distillation
Deploying these bulky architectures into medical systems to assist gastroenterologists in diagnosing the disease can be very complex, and even impractical due to the time, it takes to process the images, as well as the computational resources required to store these models. Therefore, the process of distilling knowledge from core architectures to simpler and lighter architectures will be described in this subsection. It consists on the training of large deep learning models and distilling the learnt information to teach a lighter model (with less number of parameters).
In the case of convolutional neural networks (CNNs), the knowledge distillation process consisted in scaling the logits returned by the teachers using the temperature technique, see Figure 8. A student model containing a combined total of 1,254,194 parameters was developed. Adam optimizer with a learning rate set to 1.00e-04 and the SparseCategoricalCrossEntropy loss function wase used at training. When it came to knowledge distillation, the Kullback–Leibler divergence function, represented by the formula (1), was used as the loss function. This involved configuring an alpha value of 0.8 and applying a temperature of 105 degrees in the distillation process [11].
For student training, 100 epochs and an EarlyStoping callback with a patience of five were set to monitor the validation accuracy.
In the case of ViTs, the distillation procedure was carried out in two different ways: first, by scaling the logits returned by the teachers using the temperature technique, and second, by utilising the DeiT technique [42], see Figure 9. In order to distil the 32-patched architectures in the first technique, an architecture with a total of 2,300,162 parameters was developed, while an architecture with a total of 3,442,274 parameters was built to distil the 16-patched architectures. The CNNs were distilled using the same setups.
Example of the response-base knowledge, based on Gou et al. [43]
In the second technique, an architecture with a total of 3,554,884 parameters was built to distil the architectures with 32 patches and an architecture with 3,112,516 parameters in total to do the distillation of the architectures with 16 patches. These students were configured with the AdamW optimizer with a weight decay of 0.0001 and a learning rate scaled of 6.25e-07, obtained through the formula:
The formula consisted of the quotient between the learning rate (lr), which is equal to 1.00e-05, and the base 512. The authors [42] describe the use of the 512 base as an improvement in performance. This result is multiplied by the 32 batch size.
Example of the architecture of DeiT, based on Touvron et al. [42]
The SparseCategoricalCrossentropy function was defined as the student’s loss function and as the distillation function. The students were trained for 100 epochs, using an EarlyStopping callback with a patience of five. In both distillation techniques of the ViTs, as well as in the knowledge distillation of CNNs, data augmentation was used [42, 44, 45].
3.5 Interpretability
It is important to understand in which areas of the images of the models were based to predict the type of inflammatory bowel disease, and in this sub-section, the interpretability algorithms used to interpret the output before the distillation process (teachers) and after the distillation process (students) in the CNNs and ViTs are described.
For the implementation of the CNNs, the following algorithms were selected: Grad-CAM, LIME, Shap values, RISE, and Occlusion sensitivity to interpret the teachers’ output. On the other hand, the Shap values and Grad-CAM algorithms were excluded from the interpretation of the students’ CNNs. In the ViTs implementation, the Shap values, LIME, RISE, and Occlusion sensitivity algorithms were selected to interpret the teachers’ output. In the interpretation of the students, the Shap values algorithm was excluded.
Grad-CAM and Shap values algorithms were excluded from the interpretability of the students of the CNNs, and the Shap values algorithm was not used in the interpretability from the students of the ViTs because due to the compilation of the teacher–student models in the same architecture that was not possible to get gradients of the intermediate layers.
3.6 Evaluation
The dataset was split into 70% for training, 20% for validation, and 10% for testing [18].
As previously mentioned, datasets 1 and 2 included frames extracted from videos of the HyperKvasir database (see Sect. 3.2). Two videos were used; the first contained 30 frames and presents a green square in the lower left corner. The frames extracted from this video were attached to the images in the validation set. The other video, with 34 frames, was attached to the training set.
Classification metrics were selected to evaluate the network’s and the transformer’s performance, as well as the student’s performance on the test and validation set. These include accuracy, precision, recall, F1-Score, area under curve (AUC), and inference time [46, 47]:
4 Results
After the algorithms selected for this work had been executed following the proposed methodology, the results obtained by the chosen classification metrics were collected. In this sense, this section presents through Table 5; the quantitative results obtained by the CNNs and the ViTs, as well as the results obtained by the students of each architecture.
Table 5 analyses the results obtained by the teachers and their respective students of the CNNs and ViTs. These results allow us to conclude that although the CNN and ViT teachers performed well, the inference time of the ViT teachers was greater than the inference time demonstrated by the CNN teachers. However, after the distillation of knowledge, it was more notably reduced in the ViTs than in the CNNs. Furthermore, it can be seen that in ViTs, distillation using DeiT still manages to reduce the inference time a little more when compared to the inference time of ViT distillation using temperature. However, during the DeiT distillation process, the models showed a drop in results when compared to the other distillation methods used in ViTs and even when compared to CNNs. It will also be shown in section 5 that the models during the distillation process using DeiT were unable to learn the characteristics of the two types of disease well.
Despite the good results shown by the architectures, there are some disadvantages associated with the dataset used. Firstly, we are working with very different images between the two classes to be classified, which can influence the performance of the models, but to try to overcome this problem, data augmentation was used during training to increase the generalisation capacity of the models and, as will be seen in section 5, the model is capable of predicting the disease and not the database. Another disadvantage is that we are only working with two classes (Crohn’s disease and Ulcerative colitis) instead of three classes (Crohn’s disease, Ulcerative colitis, and normal).
The absence of images of the healthy intestine could be a gap in these models in the future since they were not trained with images of the healthy intestine. However, when creating the datasets for this work, it was impossible to obtain images of the healthy intestine because we only had 14 images of the normal intestine in the CronIPI database, which would have left us with a large class imbalance. Other authors have considered Mayo 0 images as normal bowel. However, categorising these images as normal could lead to inaccuracies, as they correspond to a diagnosis of ulcerative colitis, despite the absence of disease-related lesions.
Based on the literature review carried out for this work, it was not possible to establish a comparison of results with those obtained by the authors of the papers in the literature. This is due to the fact that the authors did not use the same dataset as the one used in this study, and this causes changes in the results. Another fact is that some of the studies carried out in the literature focused on just one type of inflammatory bowel disease, so we cannot compare results from different experiments.
5 Interpretability
In addition to quantitatively evaluating the performance of the models used, it is important in this work to also qualitatively evaluate their performance through interpretability models. In order to understand which parts of the mucosa, the algorithm used as a basis to make a particular decision. Therefore, this section presents the images resulting from the interpretability of the teacher model and the interpretability of the student models, relative to the best architecture.
To make the interpretability of the models used in this work, six images were randomly selected during the creation of the datasets, referring to the classes Crohn’s disease and ulcerative colitis. Figures [10, 11, 12] present the resulting images of the interpretability of the teacher model and the student models, relative to the ViT-S/16.
The ViT-S/16 transformer is the architecture that stood out among all the models used because in quantitative terms, it achieved a good performance, and in qualitative terms. it was able to recognise more characteristics associated with each type of inflammatory bowel disease, ignoring the additional elements present in the images. Therefore, when evaluating the images collected from the model’s interpretability, it can be seen that it was able to recognise the type of inflammatory bowel disease associated with the image provided as input, by comparing the true label of the image with the label predicted by the model. It only got one image wrong during the distillation process.
It can then be seen in Fig. 10 through the interpretability of the LIME and RISE models that the ViT-S/16 architecture, using the Gaussian Blur pre-processing technique, was able to ignore the green square in the image. However, the interpretability of the occlusion sensitivity and Shap values method shows that the model already had a bias towards the additional elements in order to predict IBD. In Figs. 11 and 12, the model was able to ignore the additional elements present in the images. Based on the images, we were also able to conclude that the distillation of the transformer using DeiT was not able to learn the characteristics of the two types of IBD well when compared to the interpretability of the distillation of the transformer using temperature.
However, in the first and last ulcerative colitis images, the model was able to recognise bleeding situations, and in the last one, it was even able to identify ulcers. In the third image of Crohn’s disease, it can identify a stenosis. When comparing with Fig. 11, we see that using the distillation process through temperature, it can identify in the first image of Crohn’s disease the presence of ulcers, and in the second image, it can recognise a stenosis. In the second image of ulcerative colitis, it is also able to recognise ulcers.
6 Conclusion
It is concluded with this work that ViT models and CNNs show a good performance in the recognition of inflammatory bowel disease. It is considered that the knowledge distillation through attention in the ViTs models allowed performed to obtain a lower inference time concerning the knowledge distillation through the logits.
It is also evident in this work through the interpretability of the models that ViTs due to self-attention mechanism manage to be better than CNNs in the prediction of images that have elements external to the intestinal mucosa [48]. When analysing the quantitative and qualitative results of the ViT-S/16 is evident that the teacher model and student models can demonstrate good performance and identify Crohn’s disease lesions, ulcers, and erythema in ulcerative colitis disease, ignoring the additional elements present in the images.
This work stands out for the comparison; it offers between CNNs and ViTs architectures to perform endoscopic image classification regarding inflammatory bowel disease types. It also stands out by implementing knowledge distillation from large architectures to simpler architectures. In ViTs, two knowledge distillation approaches were implemented: (i) based on the attention of the architectures and (ii) based on the distillation of the weights through temperature to scale them.
The methods used to pre-process the images to reduce some features that might throw off the classifier when determining the type of inflammatory bowel disease are also worth mentioning. This paper further offers the interpretability of the models with the use of some model-agnostic methods, to better understand which parts of the image the architectures relied on for making a decision. Some of the results obtained from experiment 3 are documented in [49].
However, in this study, it is recognised as a limitation in the number of images used to train the ViTs because these deep learning models need a large number of images to be able to learn the information well [50]. Another limitation of this work is that the images were not collected based on the same endoscopic tool, which consequently the images are different and in the future may not demonstrate the same performance in processing colonoscopy images regarding Crohn’s disease.
As future work, it is suggested to use multi-task algorithms to make the classification of the type of lesion in the case of Crohn’s disease. As well as, the degree of severity of ulcerative colitis disease is based on the Mayo score. In addition to predicting the type of inflammatory bowel disease that the patient has, it is also important to predict the degree of the lesion in order to adapt and prescribe the best treatment. For this, ordinal techniques can be leveraged [51,52,53]. Furthermore, it is also suggested that in this validation, there should be a medical verification to ensure the veracity of the results.
Considering the difficulties currently experienced by doctors in diagnosing inflammatory bowel disease and based on the results of this study, one suggestion for the clinical use of this tool would be to help the doctor to diagnose the type of IBD the patient has. The student model would be deployed on a hospital server and, during the diagnostic examination, it would receive the images taken by the doctor, returning the class associated with the image and which characteristics linked to the type of disease were recognised. However, the use of this tool is confronted with some ethical problems, because the images collected contain personal information about the patients, which during the monitoring of this tool following the machine learning operations standard [54] ends up being somewhat exposed to third parties. Therefore, in order to guarantee the security of patient information and to comply with the GDPR [55], there are two approaches: (i) anonymise the information so that it is not clear who the images belong to and (ii) the hospital must make the patient sign a consent form for the use and processing of the data.
Data availability
Not applicable. No data were collected in the context of the development of the present work. All used datasets have been collected and made available previously by other authors. In specific, three databases of images related to Crohn’s disease and ulcerative colitis were used: HyperKvasir [23, 24]: available at https://datasets.simula.no/hyper-kvasir/. LIMUC [25]: available at https://paperswithcode.com/dataset/limuc. CrohnIPI [26,27,28,29]: available upon request to the authors
Change history
02 May 2024
A Correction to this paper has been published: https://doi.org/10.1007/s10044-024-01249-0
References
Zhang YZ (2014) Inflammatory bowel disease: pathogenesis. World J Gastroenterol 20(1):91. https://doi.org/10.3748/wjg.v20.i1.91
Ng SC, Shi HY, Hamidi N et al (2017) Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390(10114):2769–2778. https://doi.org/10.1016/S0140-6736(17)32448-0
Sairenji T, Collins KL, Evans DV (2017) An update on inflammatory bowel disease. Prim Care Clin Off Pract 44(4):673–692. https://doi.org/10.1016/j.pop.2017.07.010
Saeid Seyedian S, Nokhostin F, Dargahi Malamir M (2019) A review of the diagnosis, prevention, and treatment methods of inflammatory bowel disease. J Med Life 12(2):113–122. https://doi.org/10.25122/jml-2018-0075
Rosen MJ, Dhawan A, Saeed SA (2015) Inflammatory bowel disease in children and adolescents. JAMA Pediatr 169(11):1053. https://doi.org/10.1001/jamapediatrics.2015.1982
Huang TY, Zhan SQ, Chen PJ et al (2021) Accurate diagnosis of endoscopic mucosal healing in ulcerative colitis using deep learning and machine learning. J Chin Med Assoc 84(7):678–681. https://doi.org/10.1097/JCMA.0000000000000559
Sutton RT, Zaiane OR, Goebel R et al (2022) Artificial intelligence enabled automated diagnosis and grading of ulcerative colitis endoscopy images. Sci Rep 12(1):2748. https://doi.org/10.1038/s41598-022-06726-2
Mascarenhas MJ, Afonso J, Ribeiro T, et al (2021) Deep learning and capsule endoscopy: automatic identification and differentiation of small bowel lesions with distinct haemorrhagic potential using a convolutional neural network. p e000753, https://doi.org/10.1136/bmjgast-2021-000753
Amorim JP, Domingues I, Abreu PH, et al (2018) Interpreting deep learning models for ordinal problems. In: European symposium on artificial neural networks (ESANN), pp 373–378
Maurício J, Domingues I (2023) Deep Neural Networks to distinguish between Crohn’s disease and Ulcerative colitis. In: 11th Iberian conference on pattern recognition and image analysis (IbPRIA)
Maurício J, Domingues I (2023) Knowledge distillation of vision transformers and convolutional networks to predict inflammatory bowel disease. In: 26th Iberoamerican congress on pattern recognition
Cirrincione G, Cannata S, Cicceri G et al (2023) Transformer-based approach to melanoma detection. Sensors 23(12):5677. https://doi.org/10.3390/s23125677
Stidham RW, Liu W, Bishu S et al (2019) Performance of a deep learning model versus human reviewers in grading endoscopic disease severity of patients with ulcerative colitis. JAMA Netw Open 2(5):e193963. https://doi.org/10.1001/jamanetworkopen.2019.3963
Klang E, Barash Y, Margalit RY et al (2020) Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest Endosc 91(3):606-613.e2. https://doi.org/10.1016/j.gie.2019.11.012
Majtner T, Brodersen JB, Herp J et al (2021) A deep learning framework for autonomous detection and classification of Crohn’s disease lesions in the small bowel and colon with capsule endoscopy. Endosc Int Open 09(09):E1361–E1370. https://doi.org/10.1055/a-1507-4980
Klang E, Grinman A, Soffer S et al (2021) Automated detection of Crohn’s disease intestinal strictures on capsule endoscopy images using deep neural Networks. J Crohn’s Colitis 15(5):749–756. https://doi.org/10.1093/ecco-jcc/jjaa234
Udristoiu AL, Stefanescu D, Gruionu G, et al (2021) Deep learning algorithm for the confirmation of mucosal healing in crohn’s disease, based on confocal laser endomicroscopy images. J Gastrointest Liver Dis 30(1):59–65. https://doi.org/10.15403/jgld-3212
Chierici M, Puica N, Pozzi M et al (2022) automatically detecting crohn’s disease and ulcerative colitis from endoscopic imaging. BMC Med Inform Decis Making 22(S6):300. https://doi.org/10.1186/s12911-022-02043-w
Ruan G, Qi J, Cheng Y et al (2022) Development and validation of a deep neural network for accurate identification of endoscopic images from patients with ulcerative colitis and crohn’s disease. Front Med 9:854677. https://doi.org/10.3389/fmed.2022.854677
Wang L, Chen L, Wang X et al (2022) Development of a convolutional neural network-based colonoscopy image assessment model for differentiating crohn’s disease and ulcerative colitis. Front Med 9:789862. https://doi.org/10.3389/fmed.2022.789862
Luo X, Zhang J, Li Z et al (2022) Diagnosis of ulcerative colitis from endoscopic images based on deep learning. Biomed Signal Process Control 73:103443. https://doi.org/10.1016/j.bspc.2021.103443
Bektas B, Emre IE, Kartal E, et al (2018) Classification of mammography images by machine learning techniques. In: 2018 3rd international conference on computer science and engineering (UBMK). IEEE, Sarajevo, pp 580–585, https://doi.org/10.1109/UBMK.2018.8566380
Borgli H, Riegler M, Thambawita V et al (2019) The hyperKvasir dataset. OSF Publisher, Charlottesville
Borgli H, Thambawita V, Smedsrud PH et al (2020) HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data 7:283
Polat G, Kani HT, Ergenc I, et al (2022) Labeled images for ulcerative colitis (LIMUC) Dataset
CrohnIPI (2019) CrohnIPI. https://crohnipi.ls2n.fr/en/crohn-ipi-project/ (accessed Feb. 21, 2023)
Vallée R, Coutrot A, Normand N, et al (2021) Influence of expertise on human and machine visual attention in a medical image classification task. In: European conference on visual perception
Vallée R, Maissin A, Coutrot A, et al (2020) CrohnIPI: An endoscopic image database for the evaluation of automatic Crohn’s disease lesions recognition algorithms. In: Medical imaging: biomedical applications in molecular, structural, and functional imaging. SPIE, p 61
Vallée R, Coutrot A, Normand N, et al (2019) Accurate small bowel lesions detection in wireless capsule endoscopy images using deep recurrent attention neural network. In: IEEE 21st Int WS on Multimedia Signal Proc (MMSP)
Maurício J, Domingues I, Bernardino J (2023) Comparing vision transformers and convolutional neural networks for image classification: A literature review. Appl Sci 13(9):13095521. https://doi.org/10.3390/app13095521
Khan MN, Hasan MA, Anwar S (2021) Improving the robustness of object detection through a multi-camera-based fusion algorithm using fuzzy logic. Front Artif Intell 4:638951
Zoumpekas T, Salamó M, Puig A (2022) Effective early stopping of point cloud neural networks. In: Modeling decisions for artificial intelligence, vol 13408. Springer International Publishing, p 156–167, https://doi.org/10.1007/978-3-031-13448-7_13
Prechelt L (2012) Early stopping - but when? In: neural networks: tricks of the trade, vol 7700. Springer, Berlin-Heidelberg, p 53–67, https://doi.org/10.1007/978-3-642-35289-8_5
H. Kassani S, Hosseinzadeh Kassani P, Wesolowski M, et al (2019) Classification of histopathological biopsy images using ensemble of deep learning networks. In: arXiv preprint
Gamage C, Wijesinghe I, Chitraranjan C, et al (2019) GI-Net: anomalies classification in gastrointestinal tract through endoscopic imagery with deep learning. In: Moratuwa engineering research conference (MERCon). IEEE, Moratuwa, pp 66–71, https://doi.org/10.1109/MERCon.2019.8818929
Shahzadi I, Tang TB, Meriadeau F, et al (2018) CNN-LSTM: cascaded framework for brain tumour classification
Vankdothu R, Hameed MA, Fatima H (2022) A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput Electr Eng 101:107960
Raghu M, Unterthiner T, Kornblith S, et al (2021) Do vision transformers see like convolutional neural networks? neural information processing systems https://doi.org/10.48550/ARXIV.2108.08810
Gheflati B, Rivaz H (2021) Vision transformer for classification of breast ultrasound images. 44th annual international conference of the IEEE engineering in medicine & biology society (EMBC) https://doi.org/10.48550/ARXIV.2110.14731
Zhou HY, Lu C, Yang S, et al (2021) ConvNets versus transformers: whose visual representations are more transferable? In: IEEE/CVF international conference on computer vision workshops (ICCVW), pp 2230–2238, https://doi.org/10.1109/ICCVW54120.2021.00252
Steiner A, Kolesnikov A, Zhai X, et al (2021) How to train your ViT? data, augmentation, and regularization in vision transformers. arXiv Version Number: 2 https://doi.org/10.48550/ARXIV.2106.10270
Touvron H, Cord M, Douze M, et al (2020) Training data-efficient image transformers & distillation through attention. Arxiv https://doi.org/10.48550/ARXIV.2012.12877, publisher: arXiv Version Number: 2
Gou J, Yu B, Maybank SJ et al (2021) Knowledge distillation: a survey. Int J Comput Vis 129(6):1789–1819
Das D, Massa H, Kulkarni A, et al (2020) An empirical analysis of the impact of data augmentation on knowledge distillation. ArXiv https://doi.org/10.48550/ARXIV.2006.03810, publisher: arXiv Version Number: 2
Li W, Shao S, Liu W, et al (2023) What role does data augmentation play in knowledge distillation? In: Computer vision - ACCV 2022, vol 13842. Springer Nature Switzerland, p 507–525, https://doi.org/10.1007/978-3-031-26284-5_31, series Title: Lecture Notes in Computer Science
Chicco D, Jurman G (2020) The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21:6
Turan M, Durmus F (2022) UC-NfNet: deep learning-enabled assessment of ulcerative colitis from colonoscopy images. Med Image Anal 82:102587
Tyagi K, Pathak G, Nijhawan R, et al (2021) Detecting pneumonia using vision transformer and comparing with other techniques. In: 5th international conference on electronics, communication and aerospace technology (ICECA). IEEE, Coimbatore, pp 12–16, https://doi.org/10.1109/ICECA52323.2021.9676146
Maurício J, Domingues I (2023) Interpretability of deep neural networks to diagnose inflammatory bowel disease. In: 29th edition of the portuguese conference on pattern recognition
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. International conference on learning representations https://doi.org/10.48550/ARXIV.2010.11929
Domingues I, Cardoso JS (2014) Max-ordinal learning. IEEE Trans Neural Netw Learn Syst 25(7):1384–1389
Cardoso JS, Sousa R, Domingues I (2012) Ordinal data classification using kernel discriminant analysis: A comparison of three approaches. In: 11th international conference on machine learning and applications, pp 473–477
Marques F, Duarte H, Santos J, et al (2019) An iterative oversampling approach for ordinal classification. In: Proceedings of the 34th ACM/SIGAPP symposium on applied computing, pp 771–774
MLOps (2023) Machine learning operations. https://ml-ops.org/ (accessed Dec. 05, 2023)
GDPR (2023) General data protection regulation (GDPR). https://gdpr-info.eu/ (accessed Dec. 05, 2023)
Funding
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Contributions
These authors contributed equally to this work. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable. No data were collected in the context of the development of the present work. All used datasets have been collected and made available previously by other authors. Ethical approval was thus not needed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maurício, J., Domingues, I. Distinguishing between Crohn’s disease and ulcerative colitis using deep learning models with interpretability. Pattern Anal Applic 27, 1 (2024). https://doi.org/10.1007/s10044-023-01206-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10044-023-01206-3