Abstract
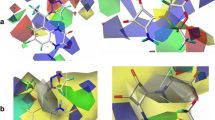
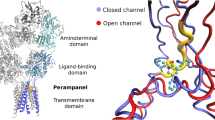
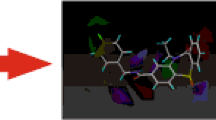
Overactivation of the N-methyl-D-aspartate receptor (NMDAR) in postsynaptic neurons leads to glutamate-related excitotoxicity in the central nervous system of mammals. We have built 3-D models of each domain for the universal screening of potential toxicants and their binding mechanisms. Our docking results show that the calculated pK i values of glycine and L-glutamate significantly increase (>1) when the NR1 and NR2A S1S2 domains are closing, respectively. Inversely, D-cycloserine (DCS) and 5,7-dichlorokynurenic acid (5,7-DCKA) do not show such a dependence on domain closure. Replica exchange molecular dynamics (REMD) confirmed 5 different conformational states of the S1S2 domain along the 308.2 K temperature trajectory. Analysis of residue fluctuations during this temperature trajectory showed that residues in loop 1, loop 2, the amino terminal domain (ATD), and the area linked to ion channel α-helices are involved in this movement. This further implicates the notion that efficacious ligands act through S1S2 lobe movement which can culminate in the opening or closing of the ion channel. We further tested this by docking hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocane (HMX) to the S1S2 domain. Our results predict that these nitramines are not efficacious and thus do not produce excitoxicity when they bind to the S1S2 domain of the NMDAR.







Similar content being viewed by others
References
Lipton SA, Nicotera P (1998) Cell Calcium 23:165–171
Choi YB, Tenneti L, Le D, Ortiz J, Bai G, Chen HSV, Lipton S (2000) Nat Neurosci 3:15–21
Meldrum B, Garthwaite J (1990) Trends Pharmacol Sci 11:379–387
Rothman SM, Olney JW (1987) Trends Neurosci 10:299–302
Dingledine R, Borges K, Bowie D, Traynelis SF (1999) Pharmacol Rev 51(1):7–61
Liu Y, Zhang J (2000) Chin Med J 113:948–956
Cull-Candy S, Brickley S, Farrant M (2001) Curr Opin Neurobiol 11:327–335
Paoletti P, Neyton J (2007) Curr Opin Pharmacol 7:39–47
Hebb DO (1949) The organization of behavior. Wiley, New York
Hebb DO (1961) Brain mechanisms and learning. Oxford University Press, London
Schorge S, Colquhoun D (2003) J Neurosci 23:1151–1158
Chatterton E, Awobuluyi M, Premkumar LS (2002) Nature 415:793–798
Furukawa H, Gouaux E (2003) EMBO J 22:2873–2885
Inanobe A, Furukawa H, Gouaux E (2005) Neuron 47:71–84
Furukawa H, Singh SK, Mancusso R, Gouaux E (2005) Nature 438:185–192
Chen HSV, Lipton SA (2005) JPET 314:961–971
Kaye SL, Sansom MSP, Biggin PC (2006) J Biol Chem 281:12736–12742
Mamonova T, Speranskiy K, Kurnikova M (2008) Proteins 73:656–671
Chohan KK, Wo ZG, Oswald RE, Sutcliffe MJ (2000) J Mol Model 6:16–25
Sutcliffe MJ, Smeeton AH, Wo ZG, Oswald RE (1998) Faraday Discuss 111:259–272
Gong P, Basu N, Scheuhammer AM, Perkins EJ (2010) Environ Sci Pollut Res Int 17:181–186
Neal AP, Worley PF, Guilarte TR (2011) Neurotoxicology 32:281–289
Nayyar T, Wu J, Hood DB (2003) Cell Mol Biol (Noisy-le-grand) 49:1357–1362
Agency for Toxic Substances and Disease Registry (1995) Toxicological profile for RDX. Agency for Toxic Substances and Disease Registry, Atlanta
Bartha R, Hsu T (1974) Soil Science 115:444–453
Jenkins TF, Walsh ME (1992) Talanta 39:419–428
Carpenter BH, Liepins R, Sickles J II, Hamilton HL, van Osdell DW (1978) SmartMedia 284:A221060
Barsotti M, Crofti G (1949) Med Lav 40:107–112
Burdette LJ, Cook LL, Dyer RS (1988) Toxicol Appl Pharmacol 92:436–444
Crouse LC, Michie MW, Major MA et al (2006) Subchronic oral toxicity of RDX in rats. Elsevier, Amsterdam, pp 619–638
Schneider NR, Bradley SR, Andersen ME (1978) Toxicol Appl Pharmacol 46:1163–1171
McCain WC, Ferguson JW (1998) Toxicol study 2340-38-95-6-1. US Army Center for Health Promotion and Preventative Medicine, Aberdeen Proving Ground
Major MA, Reddy G, Berge MA, Patzer S, Li AC, Gohdes M (2007) J Toxocol Environ Health 70:1191–1202
Quinn MJ Jr, McFarland CA, Johnson MS (2009) Toxicol Mech Methods 19:1537–6524
McFarland CA, Quinn MJ Jr, Bazar MA, Remick AK, Talent LG, Johnson MS (2008) Environ Toxicol Chem 27:1102–1111
Williams LR, Aroniadou-Anderjaska V, Qashu F, Finne H, Pidoplichko V et al (2011) Environ Health Perspect 119:357–363
Bannon DI, Dillman JF, Hable MA, Phillips CS, Perkins E (2009) J Chem Res Toxicol 22:620–625
Karp SJ, Masu M, Eki T, Ozawa K, Nakanishi SJ (1993) Biol Chem 268:3728–3733
Molina H, Horn DM, Tang N, Mathivanan S, Pandey A (2007) Proc Natl Acad Sci USA 104:2199–2204
Arnold K, Bordoli L, Kopp J, Schwede T (2006) Bioinformatics 22:195–201
Edgar RC (2004) Nucleic Acids Res 32:1792–1797
Melo F, Feytmans E (1997) J Mol Biol 267:207–222
Lüthy R, Bowie JU, Eisenberg D (1992) Nature 356:83–85
Case DA, Darden TA, Cheatham TE III et al (2008) AMBER10. University of California, San Francisco
Besler BH, Merz KM Jr, Kollman PA (1990) J Comput Chem 11:431–439
Frisch MJ, Trucks GW, Schlegel HB et al (2009) Gaussian 09, revision A.1. Gaussian Inc., Wallingford
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) J Comput Chem 19:1639–1662
Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A (2002) Bankston LA 25:474–480
Takahashi H, Shin Y, Cho SJ, Zago WM, Nakamura T, Gu Z, Ma Y, Furukawa H, Liddington R, Zhang D, Tong G, Chen HSV, Lipton SA (2007) Neuron 53:53–64
Chen PE, Geballe MT, Stansfeld PJ, Johnston AR, Yuan H, Jacob AL, Snyder JP, Traynelis SF, Wyllie DJA (2005) Mol Pharmacol 67:1470–1484
Blaise MC, Sowdhamini R, Pradhan N (2005) J Mol Model 11:489–502
Yao Y, Mayer ML (2006) J Neurosci 26:4559–4566
Acknowledgments
The authors would like to thank Jacques Reifmann and the rest of his group at The Biotechnology High Performance Computing Software Application Institute (BHSAI) for their generous allotment of time, and the Army Research Laboratory (ARL) for the use of their computational resources. This work has been done under grant number W912HZ-09-C-0026. The use of trade, product, or firm names in this report is for descriptive purposes only and does not imply endorsement by the U.S. Government. Results in this study were funded and obtained from research conducted under the Environmental Quality Technology Program of the United States Army Corps of Engineers by the US Army ERDC. Permission was granted by the Chief of Engineers to publish this information. The findings of this report are not to be construed as an official Department of the Army position unless so designated by other authorized documents.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was performed at the Environmental Laboratory of the US Army Engineer Research and Development Center (ERDC) in Vicksburg, MS 39180, USA
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
a Ramachandran plot of the NR1 S1S2 3-D model before simulation. b Ramachandran plot of the NR2A S1S2 3-D model before simulation. c Ramachandran plot of the X-ray crystal structure 1pbq chain A [14]. (DOC 102 kb)
Fig. S2
Root mean square deviation (RMSD in Å) versus time (ps) plots of a NR1 S1S2 3-D model over 4 ns and b NR2A S1S2 3-D model over 3 ns. Due to the increased flexibility observed for loops 1 and 2, the amino terminus (N), and the GT-linker regions of the NR1 S1S2 model we switched from a 2 fs time step to a 1 fs time step for the simulation of the NR2A S1S2 model. All simulations were done using the GB implicit solvent potential at 300K in the AMBER10 package. (DOC 76 kb)
Fig. S3
Backbone alignment of the NR1 open structure from a classical MD simulation (blue) and the NR1 E conformer (tan) from the REMD simulation (5.20 Å RMSD) (DOC 84 kb)
Table S1
Comparison of inhibition constants for the five different REMD conformations (DOC 49 kb)
Rights and permissions
About this article
Cite this article
Ford-Green, J., Isayev, O., Gorb, L. et al. Evaluation of natural and nitramine binding energies to 3-D models of the S1S2 domains in the N-methyl-D-aspartate receptor. J Mol Model 18, 1273–1284 (2012). https://doi.org/10.1007/s00894-011-1152-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1152-y