Abstract
Licorice is a well-known medicinal plant, containing various secondary metabolites of triterpenoid and phenolic families. The aim of this study is to evaluate the effect of salinity stress on the expression of key genes involved in the biosynthetic pathway of triterpenoids such as glycyrrhizin, betulinic acid, soyasaponins, and phytosterols in licorice root, as well as providing a phonemic platform to characterize antioxidant properties, glycyrrhizin, and total phenolic content. This study also includes measuring the gene expression level and glycyrrhizin content in leaves and roots of control plants. The studied genes included squalene synthase (SQS1 and SQS2), β-amyrin synthase (bAS), lupeol synthase (LUS), cycloartenol synthase (CAS), β-amyrin 11-oxidase (CYP88D6), and β-amyrin 24-hydroxylase (CYP93E6). Our results revealed that all of the mentioned genes were upregulated following the stress condition with different transcription rates. The highest increase (12-fold) was observed for the expression of the LUS gene, which is related to the betulinic acid pathway. Also, the highest content of glycyrrhizin was observed at 72 h post-treatment, which was consistent with the upregulated transcription levels of the glycyrrhizin pathway genes especially SQS1 and CYP88D6 at the same time. Correlation and stepwise regression analysis proved the key role of SQS1 gene in the biosynthetic pathway of glycyrrhizin. Antioxidant activity and phenolic content also were increased following stress condition. A comparison between the expression levels of SQS1 and other genes involved in the production of glycyrrhizin, phytosterols, and soyasaponins revealed a similar transcription trend, which shows the gene expression in the roots was significantly higher than the leaves. In contrast, SQS2 and LUS genes displayed a higher expression in leaf tissues. The genes related to betulinic acid biosynthetic pathway exhibited an expression rate different from other triterpenoid pathway genes, which could be observed in the leaves and roots of control plants and the roots of salt-treated plants. Furthermore, results showed that these two SQS genes have different expression rates due to different plant tissues (roots and leaves) and stress conditions. Importantly, in contrast to previous reports, we detected the glycyrrhizin in leaf tissues. This result may indicate the presence of a different genetic background in native Iranian licorice germplasm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Root and stolon of licorice (Glycyrrhiza glabra) are prevalently used in food and medicine (Bi et al. 2010; Tian et al. 2008). A number of secondary metabolites have been isolated from the licorice. The widespread use of G. glabra is attributed to the presence of two main constituents, the triterpenoid saponins and flavonoids. These compounds protect the plant against biotic and abiotic stresses and are useful natural products for human health (Nomura and Fukai 1998; Wittschier et al. 2009). Phenolic derivatives from glycyrrhiza such as liquiritigenin, isoliquiritigenin, liquritin, and isoliquiritin are the most contributors to the antioxidant potential (Martins et al. 2015). In addition, licorice produces bioactive triterpenoids such as oleanane-type triterpene saponins (glycyrrhizin and soyasaponin), lupine-type triterpenes (betulininc acid), and phytosterols, all of which together form an important class of plant secondary metabolites, displaying a wide range of biological activities (Xu et al. 2016). Glycyrrhizin is a sweet-tasting triterpene saponin in root and stolon of Glycyrrhiza. Its sweetness is estimated to be about 50 times higher than sucrose. Glycyrrhizin is a conjugate of two molecules of glucuronic acid and glycyrrhetinic acid aglycone (Hayashi 2009). It has a wide range of pharmacological, antibacterial, and antioxidant features (Sharma et al. 2013). In this regard, several studies have identified the hepato-protective (Chan et al. 2003; Jeong et al. 2002; Kimura et al. 2001), antiulcer (He et al. 2001), antiallergy (Park et al. 2004), and anticancer (Fiore et al. 2008; Salvi et al. 2003; Yoon et al. 2005) properties of this compound. Its antiviral activity against various DNA and RNA viruses (Fiore et al. 2008) has been shown in the case of HIV (De Clercq 2000) and severe acute respiratory syndrome (Cinatl et al. 2003).
Biosynthesis of secondary metabolites in plants is strongly influenced by environmental factors (Stutte 2006). As an influential factor, salt stress leads to cellular dehydration and often creates both ionic and osmotic stresses in plants (Akula and Ravishankar 2011). Studies have also demonstrated that salt stress can enhance the expression of particular genes and increase the level of secondary metabolites in some medicinal plants (Selmar 2008). It seems that the increase in the biosynthesis of secondary metabolites under stress conditions, in addition to osmotic regulation, leads to the protection of cell structures against oxidative damages (Wahid and Ghazanfar 2006).
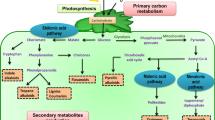
In licorice, biosynthesis of triterpenoids starts with the conversion of farnesyl diphosphate (FDP) and squalene synthase enzyme (SQS) into squalene (Lu et al. 2008). Next, squalene epoxidase oxidizes the squalene precursor to produce 2,3-oxidosqualene (Seki et al. 2011) and then oxidosqualene cyclases (OSCs) involve in cyclization of 2,3-oxidosqualene, which is an intermediate compound of triterpene saponins and phytosterols. In G. glabra, there are three key OSC enzymes: β-amyrin synthase (bAS), lupeol synthase (LUS), and cycloartenol synthase (CAS). These three enzymes are responsible for branching of triterpene saponins (glycyrrhizin and soyasaponins), betulininc acid, and phytosterols, respectively (Hayashi et al. 2000, 2001). cDNAs of SQS1 and SQS2 (Hayashi et al. 1999) along with cDNAs related to bAS, LUS, and CAS have been isolated from cultured licorice cells (Hayashi et al. 2000, 2001, 2004). In the next step, there are cytochromes P450s that play critical roles in oxidative reactions during the biosynthesis of diverse natural plant products. CYP88D6 is a cytochrome P450 that catalyzes sequential two-step oxidation reactions of β-amyrin at the C-11 position to produce 11-oxo- β-amyrin in glycyrrhizin biosynthetic pathway, whereas CYP93E is another P450 that catalyzes oxidation reaction of β-amyrin at the position of C-24 and is a key enzyme for biosynthesis of soyasaponins (Seki et al. 2008). In a study, glycyrrhizin has been found localized into stolon and woody parts of the thickened root, but not in aerial parts of the plant, whereas soyasaponins have been detected in all parts of the plants. This study showed an inverse relationship between soyasaponin and glycyrrhizin contents in licorice (Hayashi 2009).
In the present study, expression levels of SQS1, SQS2, bAS, CYP88D6, CYP93E6, CAS, and LUS genes as well as, antioxidant properties, glycyrrhizin, and total phenolic contents were measured in roots of licorice under salinity stress to obtain information on the effect of salinity on the roots.
Materials and methods
Plant material and stress treatment
Seeds of licorice (Glycyrrhiza glabra L. var. glabra, Fabaceae) were provided by Pakan-Bazr Seed Production Company (Isfahan, Iran). The seeds were disinfected using H2SO4 (98%) for 20 min and then washed with water. Next, the seeds were sown on filter papers, soaked with distilled water in Petri dishes, followed by incubation at 25 ± 2 °C (Shirazi et al. 2012). After 10 days, seedlings were transferred to hydroponic condition with Hoagland solution and incubated at 25 ± 2 °C for a photoperiod of 16 h of light (Nasrollahi et al. 2014). During this period, the aeration was conducted with an air pump and the solution was changed every 10 days. A set of 40-day-old plants representing a uniform growth trend were subjected to treatment with 150 mM NaCl in order to create moderate salt stress condition (Pan et al. 2006). The roots were harvested from seedlings subjected to different time periods of salt treatment (12, 24, 48, and 72 h) and also from seedlings under control conditions. The samples were then frozen in liquid nitrogen and were stored at − 80 °C prior to RNA extraction and other experiments. Also, parts of each root were dried at 60 °C for 48 h prior to HPLC analysis. Samples from the leaves of control plants (not their roots) were also harvested. Each of the experiment was carried out in triplicate, and biological replications were considered for each analysis.
Quantitative reverse transcription PCR
Total RNA was extracted from G. glabra roots and leaves using RNX-plus solution (Sina Clon, Iran) with slight modifications. In order to remove genomic DNA contamination, total RNA was treated with RNase-free DNase I. The first-strand cDNA synthesis was performed using 500 ng of total RNA from each sample, oligo (dT) primer, using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher).
The cDNAs were diluted 10-fold with deionized water and used as templates for quantitative reverse transcription PCR (qRT-PCR). Specific primers for SQS1, SQS2, bAS, LUS, CAS, CYP88D6, and CYP93E6 genes were designed using Oligo-7 software based on the conserved sequence of these genes in Glycyrrhiza and other Fabaceae plants (Table 1). Although SQS1 and CYP88D6 were cloned and sequenced (their data are not shown). β-Actin from Glycyrrhiza genus was also used as the housekeeping gene. The cDNAs were amplified using a reaction mixture containing 6.25 μL of 2× Green Hot Master Mix, 0.3 μM each of the forward and reverse primers (10 pmol), 1 μL of diluted cDNA template, and 4.65 μL of distilled water. PCR amplification was performed under the following conditions using BIO-RAD CFX96 Real-time PCR system: initial denaturation at 95 °C for 5 min, 45 cycles of denaturation for 20 s at 95 °C, annealing for 20 s, and extension at 72 °C for 20 s. Each sample was analyzed in triplicate and the obtained melting curves were analyzed to verify reaction specificity. In order to reveal the absence of any contamination or primer dimmers, a non-template control (NTC) reaction with each primer pair was run. To perform expression assay in leaves and roots, the expression levels of the genes were calculated by comparing the threshold cycle value (Ct) for each gene to that of the reference gene using 2−ΔCt method (Pfaffl 2001). In the case of salt stress experiment, relative gene expression was calculated using 2−ΔΔCt method (Livak and Schmittgen 2001) and the expression quantity of the samples obtained under control conditions was set as the reference value.
Chemical analysis of glycyrrhizin
To measure the glycyrrhizin content in leaves and roots of licorice, 40 mg of each dried sample was extracted with 1 mL of methanol 80% (v/v) at 60 °C for 6 h. Extract mixtures were then centrifuged at 4000 rpm for 15 min at room temperature (Hayashi et al. 1988) and, subsequently, supernatant was transferred to a new tube, followed by evaporation under dry conditions at 30 °C for 5 h.
Glycyrrhizin standard (glycyrrhizic acid ammonium salt) was purchased from Sigma Company. Prior to HPLC analysis, samples were dissolved in 1 mL of methanol and filtered through 0.45-μm filters. Separation step for glycyrrhizin was performed according to the method reported by Sabbioni et al. (2006), using an isocratic separation on reversed-phase column C18 (150 × 4.6 mm i.d.; 5 μm). The mobile phase consisted of methanol, acetonitrile, water, and glacial acetic acid (35:34:30:1 by volume). A 20-μL aliquot of each sample was analyzed by HPLC at room temperature. The flow rate was 1 mL min−1, and detection wavelength was set at 254 nm.
Total phenolic content and antioxidant properties
To measure phenolic content and antioxidant properties in control and under stress seedlings, 0.5 g of each frozen sample was extracted with 1.5 mL of ethanol 80% (v/v). Extract mixtures were then centrifuged at 10,000 rpm for 10 min at 4 °C. The supernatant was collected and stored at 4 °C before measurement. The total phenolic content was determined using a modified Folin–Ciocalteu method as described by Singleton et al. (1999). Briefly, 40 μL of the extract was mixed with 3.16 mL of distilled water, followed by addition of 200 μL of Folin–Ciocalteu reagent and allowed to stand for 8 min. Then, 600 μL of sodium carbonate solution was added. Each sample was allowed to stand for 30 min at 40 °C and the absorbance was measured at 760 nm. Results are expressed as mg gallic acid 100 g−1 FW using gallic acid calibration curve. The antioxidant properties were determined by DPPH (1,1-diphenyl-2-picrylhydrazyl) radical scavenging based on the method proposed by Du et al. (2009) with minor modifications. Briefly, 50 μL of the extract was added to 950 μL of a 6.25 × 10−5-M solution of DPPH and allowed to stand at room temperature in darkness. The absorbance of the samples was measured at 517 nm after 15 min and converted into the percentage of DPPH inhibition using the following formula.
DPPH inhibition% = [(Acontrol − Asample) / Acontrol] × 100
Statistical analysis
Cluster analysis was carried out by the Ward method based on relative gene expression data. Analysis of variance (ANOVA) and Duncan’s means comparison were used to check the significant difference in antioxidant properties, phenolic, and glycyrrhizin contents. The significance of the differences for this purpose was considered as 99% (p < 0.01). Correlation analysis between the expression of genes involved in glycyrrhizin biosynthetic pathway and glycyrrhizin content was obtained using Pearson’s correlation coefficient (r) in the case of salt treated seedlings. Stepwise regression analysis was evaluated between all relative gene expressions and the glycyrrhizin content as independent and dependent variables, respectively. Analyses were performed using SPSS (version 22.0; IBM Corp., Armonk, NY, USA). The experiment results were the mean of three replications and represented as a mean ± standard error (SE).
Results
Effect of salt stress on gene expression and glycyrrhizin content in roots
To gain an initial insight into the regulation of the key genes involved in triterpenoids biosynthetic pathway under salt stress (150 mM NaCl), relative expression rate of seven candidate genes (i.e., SQS1, SQS2, bAS, CYP88D6, CYP93E6, CAS, and LUS) was determined at different time points using qPCR (Fig. 1).
Differential expression levels of SQS1 (squalene synthase 1), SQS2 (squalene synthase 2), bAS (β-amyrin synthase), CYP88D6 (β-amyrin 11-oxidase), CYP93E6 (β-amyrin 24-hydroxylase), CAS (cycloartenol synthase), and LUS (lupeol synthase) in roots of G. glabra (40 days old plants) under salt stress condition (150 mM NaCl). Gene expression data were normalized according to β-Actin as the reference gene and relative expression were calculated toward control using 2−ΔΔCt method. Error bar is based on ± standard error
The highest level of SQS1 gene expression (2.1-fold) was observed at 72 h post-treatment. Moreover, SQS2 transcription levels significantly increased (1.9- and 2.2-fold) after stress treatment (12 and 24 h) compared to the control condition, respectively. Expression profile of bAS gene was upregulated approximately 1.6-fold at 12, 24, and 72 h of stress treatment in the beginning and at the end of the treatment period. Under stress conditions, CYP88D6 gene expression was upregulated in a monophasic pattern with a significant increase (2-fold) at 72 h and expression level of CYP93E6 was upregulated approximately 2.8-fold at 24 h post-treatment. Gene expression pattern of LUS appeared in a biphasic manner, and a significant increase was observed at 12, 24, and 72 h after the stress treatment, the highest level of expression (12-fold) identified at 12 h post-treatment. Transcriptional level of CAS gene was upregulated at 12 and 24 h after stress treatment, leading to the corresponding 1.7- and 2.3-fold increases, respectively. It appears that salt stress can effectively provoke expression activity of LUS gene. To date, quantitative analysis of the salinity stress effect on the expression pattern of LUS has not been investigated in licorice. Cluster analysis was performed based on relative gene expression under salt stress, divided into three groups; SQS1 and CYP88D6 clustered in group 1, SQS2, bAS, CYP88D6, CYP93E6, CAS in group 2, and LUS in group 3 (Fig. 2).
Cluster analysis was performed based on relative expression of SQS1 (squalene synthase 1), SQS2 (squalene synthase 2), bAS (β-amyrin synthase), CYP88D6 (β-amyrin 11-oxidase), CYP93E6 (β-amyrin 24-hydroxylase), CAS (cycloartenol synthase), and LUS (lupeol synthase) in roots of G. glabra (40 days old plants) under salt stress condition (150 mM NaCl), divided into three groups
Glycyrrhizin content in the root of licorice seedlings following salinity treatment is illustrated in Fig. 3. A significant improvement in glycyrrhizin production was observed at 72 h after salinity treatment, which is parallel to upregulation of glycyrrhizin biosynthesis genes (i.e., SQS1, bAS, and CYP88D6) at the same time. Correlation analysis between relative gene expression in glycyrrhizin pathway (SQS1, SQS2, bAS, and CYP88D6) and glycyrrhizin content revealed significant positive relationships between glycyrrhizin content and SQS1 and CYP88D6 expression levels. Also, the correlation between SQS1 and CYP88D6 showed the similarity of the expression pattern of these two genes which were previously shown by cluster analysis (Table 2). The final model in stepwise regression analysis also determined that 92% (R2) of variation in glycyrrhizin metabolism was justified with the expression level of SQS1 (Table 3), which represents the key role of this gene in the biosynthetic pathway of glycyrrhizin.
Glycyrrhizin production was profiled in roots of G. glabra (40 days old plants) under salt stress condition (150 mM NaCl) using HPLC. Bars indicate standard errors and different letters above the bars represents statistically significant differences (p < 0.01) according to Duncan’s means comparison test
Effect of salt stress on phenolic content and antioxidant properties in roots
Changes in total phenolic content due to the duration of salinity treatment in G. glabra roots are represented in Fig. 4a. A significant increase in the content of phenolic compound (40.47 mg gallic acid 100 g−1 FW) was observed at 12 h post-treatment and the lowest content was determined at 48 h post-treatment. The percentage scavenging effect of DPPH following salinity treatment is shown in Fig. 4b. The scavenging activity of DPPH was significantly increased due to prolonged stress such that the highest antioxidant activity (31%) was observed at the 48 h post-treatment.
a Total phenol content and b the percentage of DPPH inhibition activity in roots of G .glabra (40 days old plants) under salt stress condition (150 mM NaCl). Bars indicate standard errors, and different letters above the bars represent statistically significant differences (p < 0.01) according to Duncan’s means comparison test
Gene expression rate and glycyrrhizin content in roots and leaves of a control plant
Figure 5 presents the expression rates of SQS1, SQS2, bAS, CYP88D6, CYP93E6, CAS, and LUS genes in roots and leaves of 40-day-old G. glabra seedlings. As can be seen, the expression rates of SQS1 and SQS2 are turned out to be different in the roots and the leaves. SQS1 was upregulated in the root approximately 7-fold higher than that in the leaf, but expression rate of SQS2 in the leaf was 1.8-fold higher than that of the root. The highest expression levels of LUS (21-fold) and SQS2 were observed in the leaves compared to those in the roots.
Transcript levels of SQS1 (squalene synthase 1), SQS2 (squalene synthase 2), bAS (β-amyrin synthase), CYP88D6 (β-amyrin 11-oxidase), CYP93E6 (β-amyrin 24-hydroxylase), CAS (cycloartenol synthase), and LUS (lupeol synthase) in roots and leaves of G. glabra (40 days old plants). Gene expression data were normalized according to β-Actin as the reference gene, and calculated using 2−ΔCt method. Bars indicate standard errors
Glycyrrhizin content in roots and leaves was 5.256 and 0.2 μg/ g DW, respectively. Since the genes involved in the biosynthesis of glycyrrhizin (bAS and CYP88D6) were expressed in the leaves, it was expected that the production of glycyrrhizin in the leaf tissue would be also observed.
Discussion
Effect of salt stress on gene expression of triterpenoids pathway in roots
Cluster analysis of relative gene expression was classified the genes into three groups; SQS1 and CYP88D6 genes associated with glycyrrhizin pathway were classified in group 1; the genes related to soyasaponins and phytosterols were in group 2; and a single gene related to the biosynthetic pathway of betulinic acid was placed in a separate group 3. Given these results, it appears that salt stress can provoke the expression activity of SQS1 and SQS2 genes in different patterns. Lee et al. (2004) demonstrated SQS upregulation in hairy root cultures of Panax ginseng under methyl jasmonate (MeJA) treatment. In addition, Hayashi et al. (2004) reported that levels of both SQS transcripts were increased by the addition of MeJA to the cell culture of licorice. Squalene synthases are a multi-gene family, of which SQS1 and SQS2 have been identified in the triterpenoid biosynthetic pathway of licorice so far (Hayashi et al. 1999). SQS function takes place at the start of the triterpenoid biosynthetic pathway (Lu et al. 2008); hence, it is expected that expression of downstream genes involved in the triterpenoid pathway could be affected by an increase in transcription levels of these two upstream genes. The bAS gene is located at the downstream of SQS and separates the biosynthetic pathway of glycyrrhizin and soyasaponin from betulinic acid and phytosterol. The bAS enzyme produces β-amyrin, which is an intermediate compound in soyasaponin and glycyrrhizin pathways. Previous studies have reported that transcriptional expression level of bAS in licorice cell culture increased with MeJA and decreased with yeast extract treatments (Hayashi et al. 2004). Nasrollahi et al. (2014) also demonstrated the upregulation of bAS in the seedlings of G. glabra under drought stress. In the present study, expression levels of bAS gene were found to be associated with expression patterns of SQS1 and SQS2 genes. Interestingly, the expression level of SQS2 may be more effective in expressing the bAS gene.
CYP88D6 enzyme creates a detour of glycyrrhizin production pathway from that of soyasaponins. CYP88D6-mediated oxidation of β-amyrin at the C-11 and C-30 positions leads to glycyrrhizin production. However, the oxidation reaction is blocked at these positions in cell culture condition; β-amyrin is oxidized by CYP93E6 at C-22 and C-24 positions, which leads to the formation of soyasapogenol B (Seki et al. 2008). In an RNA-sequencing project of G. uralensis, the highest expression level of CYP88D6 gene was observed in licorice strains collected during the summer (Ramilowski et al. 2013). So far, expression patterns of CYP88D6 and CYP93E6 genes have not yet been studied under stress conditions. In the present study, expression levels of these genes were found to be associated with SQS1 and SQS2 expressions, respectively. Also, correlation analysis revealed a significant positive relationship between the genes related to glycyrrhizin pathway (SQS1 and CYP88D6). In the case of CYP88D6, an increased level of expression was observed at 72 h after stress treatment, which was similar to the over-expression pattern of SQS1 and bAS at 72 h post-treatment. The expression level of CYP93E6 gene was also increased by approximately 2.8-fold at 24 h of salinity stress compared to the control, which was similar to the transcription patterns of SQS2 and bAS. Both CYP88D6 and CYP93E6 expression levels were decreased at 48 h after the salinity treatment, similar to the expression levels of SQS1, SQS2, and bAS. Contents of glycyrrhizin and soyasaponin were measured in different organs of licorice using liquid and gas chromatography methods (i.e., HPLC and GC, respectively). The results showed that there was a negative correlation between glycyrrhizin and sayasaponin contents in different organs (Hayashi et al. 1988, 1993, 2004). Clustering results of this study also confirmed that salt stress has a diverse effect on transcription levels of the genes involved in the production of these two competitive metabolites. In addition to triterpenoids, higher plants usually produce phytosterols, which play a structural role in the membrane system (Hayashi et al. 2004). CAS is an oxiosqulalene cyclase enzyme that accelerates the formation of cycloartenol from 2,3-oxidosqualene. Cycloartenol is the first precursor in the biosynthetic pathway of phytosterols in higher plants. In this study, the expression level of CAS was similar to the expression pattern of SQS2 and both were upregulated by salt stress. According to the results of Nasrollahi et al. (2014), transcript levels of CAS gene did not exhibit significant differences in either control or drought stress samples. Also, Hayashi et al. (2004) observed no differences in gene expression levels of CAS gene in cell cultures treated with MeJA and gibberellic acid (GA3); instead, a difference was observed in its expression in the root and stolon tissues obtained from G. uralensis (Tamura et al. 2016). LUS gene expression was significantly increased under salinity stress; hence, salinity might lead to an increase in betulinic acid levels. Transcripts of the LUS gene could be monitored in parts of the plant with higher betulinic acid content (Hayashi et al. 2004). In the research conducted by Nasrollahi et al. (2014), any transcripts of the LUS were determined in 8-day-old seedlings and stolons of licorice in control and drought treatment conditions using semi-quantitative RT-PCR. Cell cultures treated with MeJA and GA3 showed the suppression of LUS by MeJA, whereas the transcription level was remained unchanged following GA3 treatment (Hayashi et al. 2004). Tamura et al. (2016) observed betulinic acid only in the tissue cultures of stolon, but not in the normal roots; so, the expression of this gene was also minimal in control roots of licorice.
Results of this research confirmed that the expression profiles of the triterpenoid biosynthesis genes were upregulated under the salt stress conditions. Furthermore, it seems that different time periods of salt stress exhibited different gene expression profiles for each of triterpenoid pathway genes. Salt treatment caused the greatest effect on LUS gene expression, which was related to the betulinic acid pathway.
Effect of salt stress on glycyrrhizin content in roots
Significant positive corrections were identified between glycyrrhizin content and SQS1 and CYP88D6 gene expressions, individually. These data showed the great impact of these two genes on the glycyrrhizin biosynthetic pathway. In the case of a research on metabolic engineering of glycyrrhizin biosynthetic pathways including SQS1 and CYP88D6, an increased amount of glycyrrhizin was observed in hairy root cultures of Glycyrrhiza plants that confirmed the results of this study (Lu et al. 2008; Shirazi et al. 2018). Production pattern of glycyrrhizin was more similar to the expression pattern of SQS1 gene so that the stepwise regression analysis also confirmed that 92% of the change in glycyrrhizin content is associated with SQS1. Consequently, it was hypothesized that SQS1 activity provides the bulk of the precursors required for biosynthesis of glycyrrhizin. However, all of the genes involved in the biosynthetic pathway, from the beginning to the end, have important roles in the production of glycyrrhizin. Shabani et al. (2010) reported an increase in production levels of glycyrrhizin followed by increased expression of SQS genes. Also, Nasrollahi et al. (2014) demonstrated that an improvement in glycyrrhizin production occurred following a severe drought stress, which is consistent with the results of this study that glycyrrhizin has increased by prolonged stress. Glycyrrhizin and its hydrolyzed metabolites such as 18 β-glycyrrhetinic acid possess antioxidant properties, which lead to the reduction of oxidative damages (Kim and Lee 2008). It has been revealed that glycyrrhizin can reduce the harmful effects of stress on plant defense systems and also plays an important role in adaptability to stress conditions. In fact, the main role of secondary metabolites in plants is to protect them against adverse conditions. Further studies, hence, should be conducted to assess the effects of long-term and more intensive salinity treatments.
Phenolic content and antioxidant properties
In the present study, total phenolic compounds were increased at 12 h of salinity stress in roots. Plant secondary metabolites especially phenolic and flavonoid compound act as antioxidants and has a radical scavenging ability. The redox attributes of phenolic metabolites are facilitated by their hydroxyl groups (Soobrattee et al. 2005). Environmental stress conditions could increase a variety of reactive oxygen species, so that these free radicals caused the severe oxidative damage and subsequent tissue injury in plants (Saeed et al. 2012). The physiological stresses can considerably increase the level of phenylalanine ammonia lyase (PAL), an important enzyme involved in phenolic biosynthesis; thus, more phenolic compounds accumulated following the stress conditions (Morello et al. 2005).
Using the DPPH radical scavenging method, we demonstrated that the antioxidant properties were significantly increased in different periods of salt treatment. This result can be attributed to an increase in the expression of genes and the content of secondary metabolites such as triterpenoid and phenolic compounds. Antioxidant activity is influenced by several factors and cannot be completely described by only a single reaction system. Thus, multiple assays need to evaluate antioxidant activity and may provide exclusive information on their multiple abilities to scavenge different radicals (Karami et al. 2013).
Expression rates of the genes associated with a triterpenoid pathway in roots and leaves
SQS1 gene expression rate is very similar to those of phytosterol, soyasaponins, and glycyrrhizin pathway genes such as CAS, bAS, CYP88D6, and CYP93E6, representing higher levels of expression in the roots in comparison with the leaves (1.7-, 5.6-, 2.7-, and 5.5-fold, respectively). Expression levels of SQS1 and SQS2 genes have been determined in roots, stems, leaves, flowers, and young seeds of soybean by real-time PCR and demonstrated that both genes were differentially expressed in all the studied tissues. However, SQS1 gene was highly transcribed in roots in comparison with the other tissues (Nguyen et al. 2013). Three SQS genes have been identified in Ginseng, which was differentially expressed. Although all the three SQS enzymes are involved in squalene production, SQS1 enzyme was expressed in all the organs, whereas SQS2 and SQS3 exhibited a tissue-dependent expression (Kim et al. 2011). Overall, a higher ratio of SQS gene expression in roots of Fabaceae family was found to be consistent with the higher accumulation levels of phytosterols and saponins in roots of these plants (Akamine et al. 2003; Suzuki et al. 2002). Transcription of LUS was found to be very low in comparison with the other genes in the control plant, and its expression level was not similar to those of the two other oxidosqualene cyclases. These findings presumably suggest that betulinic acid production pathway is different from soyasaponins, phytosterols, and glycyrrhizin biosynthetic pathways.
Expression levels of LUS, CAS, bAS, CYP88D6, and CYP93E3 in root and stolon of G. uralensis demonstrated different expression rates. Moreover, transcription levels of CAS, bAS, CYP88D6, and CYP93E3 genes in roots were higher compared to the stolons. However, LUS gene represented higher and lower expression levels in stolon and root tissues, respectively (Tamura et al. 2016). Studies on the expression of CAS gene using semi-quantitative RT-PCR in root nodules, thickened roots, and cell cultures revealed that gene expression levels were constant in all the tissues (Hayashi et al. 2004). In addition, CAS expression displayed no changes during different studied months. In semi-quantitative expression analysis, there was no evidence for the expression of bAS and CYP88D6 genes in leaves (Seki et al. 2008, 2011). In the present study, bAS and CYP88D6 genes were expressed in leaf tissues, but the expression of both genes in roots was higher than that in leaves. However, the transcript levels of almost mentioned genes were low, with the highest level being related to bAS in root tissues, which is associated with higher levels of soyasaponin and glycyrrhizin in different parts of the licorice root.
Glycyrrhizin production in roots and leaves
Metabolic evaluation of different parts of licorice using HPLC and GC exhibited that glycyrrhizin is located in thickened roots of the plants. In aerial parts, rootlets, and root nodules, glycyrrhizin was not observed, however (Hayashi 2009). Unlike previous researches, in the present study, glycyrrhizin production was observed in leaf tissues. Nevertheless, its content was less than that of the root, which is the main organ of glycyrrhizin production. Genetic diversity among different species and the variety of environments are the main factors affecting the biosynthesis of glycyrrhizin (Kojoma et al. 2011; Zhang and Ye 2009). Overall, for studies regarding the biosynthetic pathway of glycyrrhizin, it is suggested using the variety that produces more glycyrrhizin (Kojoma et al. 2011; Zhang and Ye 2009). Glycyrrhizin production in leaves of the 40-day-old seedlings is considered as a promising source for acquiring glycyrrhizin metabolite from the leaves of licorice.
Conclusions
Salinity stress had a positive effect on the expression of key genes involved in all three biosynthetic pathways of phytosterols, oleananes, and lupine saponins in roots. However, its effect on each of these genes varied by different exposure times. Therefore, gene expressions of phytosterol, betulinic acid, and soyasaponins pathways increased at the beginning of the treatment period, while gene expressions of glycyrrhizin and betulinic acid increased at the end of the treatment period. As expected, antioxidant activity, glycyrrhizin content, and total phenolic content were increased followed by salinity stress. SQS1, bAS, CAS, CYP88D6, and CYP93E6 represented higher expression levels in the roots of the control plant, but SQS2 and LUS were expressed more in the leaf tissue. If the increase and decrease in the expression of downstream genes and the production of enzymes occur with the same trend, it is expected that production of glycyrrhizin, soyasaponin, and phytosterol in roots would be probably higher than that of the leaves while betulinic acid would be higher in the leaves. In this regard, measuring the metabolites in both leaf and root tissues are required to prove these findings. The results obtained from the expression analysis of the genes under control condition (leaves and roots) and salinity stress treatment (roots) revealed the different expression profiles of the betulinic acid biosynthetic pathway from other triterpenoids pathways. These results also demonstrated that SQS gene family might have different roles in triterpenoids biosynthetic pathways in different parts of the plants (leaves and roots) and different stress conditions in roots. Regarding the expression of glycyrrhizin pathway genes in the leaves, glycyrrhizin metabolite was also produced in the leaf tissue, which could indicate a different genetic background of native Iranian licorice. In addition, treatments with higher salinity concentrations are suggested for analyzing the expression of genes along with the measurement of their metabolites in long-term following the salt treatment. Moreover, the glycyrrhizin contents of perennial Iranian licorices collected from different parts of the country could be measured. If these plants turn out to have high levels of glycyrrhizin, then plant leaves can also be used to extract this metabolite, which facilitates the extraction process and could be more cost-effective compared to extraction from stolon or root.
Abbreviations
- bAS:
-
β-Amyrin synthase
- CAS:
-
Cycloartenol synthase
- CYP88D6:
-
β-Amyrin 11-oxidase
- CYP93E6:
-
β-Amyrin 24-hydroxylase
- FDP:
-
Farnesyl diphosphate
- GA3:
-
Gibberellic acid
- HPLC:
-
High-performance liquid chromatography
- LUS:
-
Lupeol synthase
- MeJA:
-
Methyl jasmonate
- OSCs:
-
Oxidosqualene cyclases
- QRT-PCR:
-
Quantitative reverse transcription PCR
- SQS:
-
Squalene synthase
References
Akamine S, Nakamori K, Chechetka SA, Banba MY, Umehara H, Kouchi K, Hata S (2003) cDNA cloning, mRNA expression, and mutational analysis of the squalene synthase gene of Lotus japonicus. Biochim Biophys Acta 1626:97–101
Akula R, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6:1720–1731
Bi W, Tian M, Row KH (2010) Solid-phase extraction of liquiritin and glycyrrhizin from licorice using porous alkyl-pyridinium polymer sorbent. Phytochem Anal 21:496–501
Chan HT, Chan C, Ho JW (2003) Inhibition of glycyrrhizic acid on aflatoxin B 1-induced cytotoxicity in hepatoma cells. Toxicology 188:211–217
Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr H (2003) Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361:2045–2046
De Clercq E (2000) Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Med Res Rev 20:323–349
Du G, Li M, Ma F, Liang D (2009) Antioxidant capacity and the relationship with polyphenol and vitamin C in Actinidia fruits. Food Chem 113(2):557–562
Fiore C, Eisenhut M, Krausse R, Ragazzi E, Pellati D, Armanini D, Bielenberg J (2008) Antiviral effects of Glycyrrhiza species. Phytother Res 22:141–148
Hayashi H (2009) Molecular biology of secondary metabolism: case study for Glycyrrhiza plants. In: Recent Advances in Plant Biotechnology. Springer, New York/Dordrecht, pp 89–103
Hayashi H, Fukui H, Tabata M (1988) Examination of triterpenoids produced by callus and cell suspension cultures of Glycyrrhiza glabra. Plant Cell Rep 7:508–511
Hayashi H, Fukui H, Tabata M (1993) Distribution pattern of saponins in different organs of Glycyrrhiza glabra. Planta Med 59:351–353
Hayashi H, Hiraoka N, Ikeshiro Y, Kushiro T, Morita M, Shibuya M, Ebizuka Y (2000) Molecular cloning and characterization of a cDNA for Glycyrrhiza glabra cycloartenol synthase. Biol Pharm Bull 23:231–234
Hayashi H, Hirota A, Hiraoka N, Ikeshiro Y (1999) Molecular cloning and characterization of two cDNAs for Glycyrrhiza glabra squalene synthase. Biol Pharm Bull 22:947–950
Hayashi H, Huang P, Kirakosyan A, Inoue K, Hiraoka N, Ikeshiro Y, Kushiro T, Shibuya M, Ebizuka Y (2001) Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol Pharm Bull 24:912–916
Hayashi H, Huang P, Takada S, Obinata M, Inoue K, Shibuya M, Ebizuka Y (2004) Differential expression of three oxidosqualene cyclase mRNAs in Glycyrrhiza glabra. Biol Pharm Bull 27:1086–1092
He JX, Akao T, Nishino T, Tani T (2001) The influence of commonly prescribed synthetic drugs for peptic ulcer on the pharmacokinetic fate of glycyrrhizin from Shaoyao-Gancao-Tang. Biol Pharm Bull 24:1395–1399
Jeong HG, You HJ, Park SJ, Moon AR, Chung YC, Kang SK, Chun HK (2002) Hepatoprotective effects of 18β-glycyrrhetinic acid on carbon tetrachloride-induced liver injury: inhibition of cytochrome P450 2E1 expression. Pharmacol Res 46:221–227
Karami Z, Mirzaei H, Emam-Djomeh Z, Mahoonak AS, Khomeiri M (2013) Effect of harvest time on antioxidant activity of Glycyrrhiza glabra root extract and evaluation of its antibacterial activity. Int Food Res J 20(5):2951–2957
Kim TD, Han JY, Huh GH, Choi YE (2011) Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol 52:125–137
Kimura M, Inoue H, Hirabayashi K, Natsume H, Ogihara M (2001) Glycyrrhizin and some analogues induce growth of primary cultured adult rat hepatocytes via epidermal growth factor receptors. Eur J Pharmacol 431:151–161
Kim YJ, Lee CS (2008) Glycyrrhizin attenuates MPTP neurotoxicity in mouse and MPP+-induced cell death in PC12 cells. Korean J Physiol Pharmacol 12:65–71
Kojoma M, Hayashi S, Shibata T, Yamamoto Y, Sekizaki H (2011) Variation of glycyrrhizin and liquiritin contents within a population of 5-year-old licorice (Glycyrrhiza uralensis) plants cultivated under the same conditions. Biol Pharm Bull 34:1334–1337
Lee MH, Jeong JH, Seo JW, Shin CG, Kim YS, In JG, Yang DC, Yi JS, Choi YE (2004) Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol 45:976–984
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lu HY, Liu JM, Zhang HC, Yin T, Gao SL (2008) Ri-mediated transformation of Glycyrrhiza uralensis with a squalene synthase gene (GuSQS1) for production of glycyrrhizin. Plant Mol Biol Report 26:1–11
Martins N, Barros L, Duenas M, Santos-Buelga C, Ferreira IC (2015) Characterization of phenolic compounds and antioxidant properties of Glycyrrhiza glabra L. rhizomes and roots. RSC Adv 5:26991–26997
Morello JR, Romero MP, RamoT MMJ (2005) Evaluation of L-phenylalanine ammonia-lyaseactivity and phenolic profile in olive drupe (Oleaeuropaea L.) from fruit setting period to harvestingtime. Plant Sci 168:65–72
Nasrollahi V, Mirzaie-asl A, Piri K, Nazeri S, Mehrabi R (2014) The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 103:32–37
Nguyen HT, Neelakadan AK, Quach TN, Valliyodan B, Kumar R, Zhang Z, Nguyen HT (2013) Molecular characterization of Glycine max squalene synthase genes in seed phytosterol biosynthesis. Plant Physiol Biochem 73:23–32
Nomura T, Fukai T (1998) Phenolic constituents of licorice (Glycyrrhiza species). In: Herz W, Kirby GW, Moore RE, Steglich W, Tamm C (eds) Fortschritte der Chemie Organischer Naturstoffe. Springer Verlag
Pan Y, Wu LJ, Yu ZL (2006) Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul 2:157–165
Park SH, Yoon HK, Han MJ, Kim DH (2004) Anti-allergic activity of 18β-glycyrrhetinic acid-3-O-β-D-glucuronide. Arch Pharm Res 27:57–60
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45–e445
Ramilowski JA, Sawai S, Seki H, Mochida K, Yoshida T, Sakurai T, Muranaka T, Saito K, Daub CO (2013) Glycyrrhiza uralensis transcriptome landscape and study of phytochemicals. Plant Cell Physiol 54:697–710
Sabbioni C, Ferranti A, Bugamelli F, Forti GC, Raggi MA (2006) Simultaneous HPLC analysis, with isocratic elution, of glycyrrhizin and glycyrrhetic acid in liquorice roots and confectionery products. Phytochem Anal 17:25–31
Saeed N, Khan MR, Shabbir M (2012) Antioxidant activity, total phenolic and total flavonoid contents of whole plant extracts Torilis leptophylla L. BMC Complement Altern Med 16:212–221
Salvi M, Fiore C, Armanini D, Toninello A (2003) Glycyrrhetinic acid-induced permeability transition in rat liver mitochondria. Biochem Pharmacol 66:2375–2379
Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci 105:14204–14209
Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23:4112–4123
Selmar D (2008) Potential of salt and drought stress to increase pharmaceutical significant secondary compounds in plants. Landbauforsch Volk 58:139–144
Shabani L, Ehsanpour A, Esmaeili A (2010) Assessment of squalene synthase and beta-amyrin synthase gene expression in licorice roots treated with methyl jasmonate and salicylic acid using real-time qPCR. Russ J Plant Physiol 57:480–484
Sharma V, Agrawal RC, Pandey S (2013) Phytochemical screening and determination of anti-bacterial and anti-oxidant potential of Glycyrrhiza glabra root extracts. J Environ Res Develop 7:1552–1558
Shirazi Z, Aalami A, Tohidfar M, Sohani MM (2018) Metabolic engineering of glycyrrhizin pathway by over-expression of beta-amyrin 11-oxidase in transgenic roots of Glycyrrhiza glabra. Mol Biotechnol 60:412–419
Shirazi Z, Piri K, Mirzaie-asl A, Hasanloo T (2012) Glycyrrhizin and isoliquiritigenin production by hairy root culture of Glycyrrhiza glabra. J Med Plant Res 6:4640–4646
Singleton VL, Orthofer R, Lamuela-Raventós RM (1999) Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Methods Enzymol 299:152–178
Soobrattee MA, Neergheen VS, Luximon-Ramma A, Aruoma OI, Bahorun OT (2005) Phenolics as potential antioxidant therapeutic agents: mechanism and actions. Mutat Res 579: 200–213
Stutte GW (2006) Process and product: recirculating hydroponics and bioactive compounds in a controlled environment. HortScience J 41:526–530
Suzuki H, Achnine L, Xu R, Matsuda S, Dixon RA (2002) A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J 32:1033–1048
Tamura K, Seki H, Suzuki H, Kojoma M, Saito K, Muranaka T (2016) CYP716A179 functions as a triterpene C-28 oxidase in tissue-cultured stolons of Glycyrrhiza uralensis. Plant Cell Rep 36:437–445
Tian M, Yan H, Row KH (2008) Extraction of glycyrrhizic acid and glabridin from licorice. Int J Mol Sci 9:571–577
Wahid A, Ghazanfar A (2006) Possible involvement of some secondary metabolites in salt tolerance of sugar cane. J Plant Physiol 163:723–730
Wittschier N, Faller G, Hensel A (2009) Aqueous extracts and polysaccharides from liquorice roots (Glycyrrhiza glabra L.) inhibit adhesion of Helicobacter pylori to human gastric mucosa. J Ethnopharmacol 125:218–223
Xu G, Cai W, GaoW LC (2016) A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol 212:123–135
Yoon G, Do Jung Y, Cheon SH (2005) Cytotoxic allyl retrochalcone from the roots of Glycyrrhiza inflata. Chem Pharm Bull 53:694–695
Zhang Q, Ye M (2009) Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J Chromatogr A 1216:1954–1969
Acknowledgments
The authors would like to express their sincere gratitude to the Department of Plant Biotechnology, University of Guilan, Rasht, Iran.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Peter Nick
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shirazi, Z., Aalami, A., Tohidfar, M. et al. Triterpenoid gene expression and phytochemical content in Iranian licorice under salinity stress. Protoplasma 256, 827–837 (2019). https://doi.org/10.1007/s00709-018-01340-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-01340-4