Abstract
The ligand H2L (N-(N,N-diethylaminothiocarbonyl)benzimidoylchloride-2-aminoacetophenone-N-methylthiosemicarbazone) reacts with ruthenium(II) building blocks [RuHCl(CO)(EPh3)3] (E = P or As) to form new complexes [Ru(1,1-DT)(Cl2)(CO)-(EPh3)2] (E = P or As; 1,1-DT = 1,1-diethylthiourea). The ligand H2L in these reactions undergoes C=N bond break and coordinates through sulfur atom of C=S group. Analytical and spectral (IR, UV–Vis, NMR, ESI-MS) methods were used to characterize the compounds. A distorted octahedral geometry for complexes has been furnished by X-ray crystallography, which confirmed the coordination mode of the ligand with metal precursor. The binding affinity and mode of binding of the complex towards some important biomolecules such as calf thymus DNA and bovine serum albumin protein were determined using absorption and emission spectra and found intercalative binding with calf thymus DNA and static interaction with bovine serum albumin. The in vitro cytotoxic activity of complex was assessed using human cervical cancer (HeLa), human hepatocellular carcinoma (HepG2), and normal Vero cells. Furthermore, the complex was found to possess significant enzyme mimic catalytic activity in oxidation and hydrolysis reactions.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, attention has been given for metallic drug–DNA interaction in inorganic pharmaceutical research. Due to the tunable coordination properties of the inorganic drugs, the interaction of transition metal complexes with molecular target DNA is of interest for understanding their cytotoxic activities [1,2,3]. These compounds can bind with DNA duplex through non-covalent interactions, viz intercalation between the bases, surface groove-binding, and electrostatic attraction with anionic sugar–phosphate backbones [4]. Based on the interaction of biomolecules and metal complexes, search for new metal-based drugs for cancer treatment has led to an increasing significance for the cytotoxic properties of metal complexes and their mechanisms of action [5, 6]. One of the great accomplishments in the domain of medicinal inorganic chemistry is the introduction of cis-diamminedichloroplatinum(II) (cis-platin) and its successors in the treatment of cancer. However, few of their severe side effects such as renal toxicity, neurotoxicity, myelosuppression, immunosuppression, severe nausea, vomiting, and ototoxicity impelled inorganic chemists to explore new anticancer drugs based on transition metal complexes [7]. Among the various transition metal complexes scrutinized, the ruthenium complexes are the most favorable alternatives in this regard due to their ease of synthesis, advanced chemical and air stability, solubility, rich redox chemistry, structural diversity, and kinetic aspects akin to platinum [8, 9]. Presently, few ruthenium complexes were entered in clinical trials as anticancer agents [10].
On the other hand, over the past few decades, attention has been made to artificial enzymes, which mimic the structural and functional aspects of naturally occurring enzymes, for utilizing oxidation of organic substrates in the arena of industrial and synthetic processes [11]. In this direction, many catalysts having bio-mimicking activity for various enzymes have been synthesized by chemists. Such artificial enzymes have the same catalytic function, but they are more stable, cost-efficient, and structurally less complex than their natural counterparts. One of the prominent natural catecholase enzymes, a less well-known member of the type-III copper proteins that originate in plant tissues and crustaceans, plays a key role in the oxidation of a wide range of o-diphenols (catechol) to the corresponding o-quinones [12, 13], a process known as catecholase activity. Similarly, hydrolytic reactions, i.e., hydrolysis of phosphodiester bond (phosphodiester cleavage), a process called phosphatase activity, also involves enzyme catalysis. Many research groups are working towards synthesis of enzymes with different materials including metal complexes to imitate the biological functions of above natural enzymes. Recently, our group has reported metal complexes with various ligands [14], which mimic catecholase and phosphatase activities.
Generally, the nature of ligands and chelation decides the properties of the metal complexes. Nowadays, interest is being given to develop thiourea-based ligands that support highly reactive transition metal complexes. N,N-[(Dialkylamino)(thiocarbonyl)]benzamidine has been observed as a diverse selection of multifunctional thiourea scaffolds that can support metal ions by providing a wide variety of different steric and electronic environments [15, 16]. A new class of tridentate thiocarbanoylbenzamidines containing an additional thiosemicarbazone moiety and their rhenium complexes have been reported recently together with the biological activity of the compounds [17].
Motivated on the above contemplations and with the aim to study transition metal complex interaction on biomolecules, we have synthesized ruthenium(II) complexes containing thiourea ligands (Scheme 1). The interaction studies of ruthenium(II) complex have been performed on selected biomolecules such as calf thymus DNA (CT-DNA) and bovine serum albumin (BSA) to determine the binding mode. The cytotoxic as well as enzyme mimic activities of the complex were also done to understand the biological reactivity preferences.

Results and discussion
Synthesis and characterization
The ligand H2L was obtained by simple condensation using our previously reported methodology [18] with crystalline purity and sufficient yield to use without further purification for the complex synthesis. Successively, ruthenium(II) complexes 1, 2 were synthesized in good yields by the reaction of H2L and stable prefabricated ruthenium(II) precursors [RuHCl(CO)(EPh3)3)] (E = P or As) in CH3OH/CHCl3 solvent mixture as shown in Scheme 1. Interestingly, the ligand undergoes hydrolytic C=N bond cleavage and coordinates monodentate mode via sulfur of C=S group with ruthenium metal.
The new complexes are crystalline solids, non-hygroscopic, air-stable in solution and solid state at room temperature. The complexes were characterized satisfactorily by analytical, spectral (IR, UV–Vis, NMR, and ESI-MS) as well as single crystal X-ray crystallography studies.
Spectroscopic analysis
The typical IR bands of the new ruthenium(II) complexes were compared with those of the free ligand to find the coordination mode of ligand in complexes. The spectrum of ligand shows three peaks in the range of 3236–3302/cm due to νNH groups, whereas in the spectra of complexes the νNH peaks disappeared indicates deprotonation of these groups in ligand on complexation. However, in complexes 1 and 2 a new broad band was observed in the region at 3379–3381/cm specified to new NH2 group formed due to C=N bond break in the benzamidine ligands [19]. A sharp band appeared range at 820–843/cm in the spectrum of ligand was assigned to ν C=S group which shifted to lower wave number in complexes 1 and 2 upon complex formation. The shift of ν C=S by 20–30 unit in complexes indicates that the ligand is coordinated to the ruthenium(II) center via the sulfur atom. In addition, the ruthenium(II) complexes show strong vibrations in the anticipated region conform the presence of triphenylphosphine and triphenylarsine. The electronic spectra of ligand and complexes show two–four bands in the region 237–443 nm. The bands in the 237–272 nm regions can be assigned to π → π* and n → π* transitions [20]. The absorption maxima located in the 332–339 nm range were assigned to S(pπ) → M(dπ) (M = Ru2+) LMCT transition for complexes 1 and 2. Furthermore, a broad band at 438–443 nm region in the spectra of complexes have been assigned to d–d transition band of spin-paired d6 species with a distorted octahedral structure.
The 1H NMR spectrum of ligand H2L shows three different NH resonances at 8.10, 8.47, and 12.72 ppm. But in 1 and 2, the peaks for NH groups were completely disappeared and a new peak appeared around 8.17–8.21 ppm for –NH2 due to ligand C=N bond cleavage. The four well separated multiplet resonances at 3.52–3.94 ppm have been assigned to two methylene groups in ligand and its complexes 1 and 2. Furthermore, appearance of two triplet signals region at 1.15–1.62 ppm were assigned to N(CH2CH3)2 methyl groups in the spectra of ligand and its complexes [21, 22]. In addition, signals for aromatic protons appear in the expected region. In the 13C NMR spectrum of uncoordinated ligand H2L, the C=N and C=S signals appear around 149.36–155.41 and 187.08–189.61 ppm, respectively [23]. In the spectra of Ru(II) complexes, the signals of the C=N carbon disappeared and C=S carbon signals shifted to upfield region (169.19–171.69 ppm). These are consistent with the sulfur donor coordination for complexes 1 and 2 and break of C=N bond in benzamidine ligands. The carbonyl carbon resonates at 203.01–204.12 ppm, was comparable with the literature reports. In addition, peaks for CH2 and CH3 carbon were observed region at 13.21–32.61 ppm. Furthermore, the aromatic carbons in the complexes exhibit peaks in the region of 127.62–138.83 ppm. The 31P NMR spectrum of complex 1 show a sharp singlet at 34.83 ppm indicates the coordination of two magnetically equivalent triphenylphosphine co ligands in trans position [24]. The mass spectral analysis of complexes 1, 2 reveals a signal that corresponds to a molecular ion at m/z = 821.32, 909.22, respectively, which is assigned to [M−Cl]+ with an experimental isotope pattern that matches the calculated values.
X-ray crystallographic analysis
X-ray crystallographic analysis of 1 and 2 also confirms the metal-induced hydrolytic C=N bond break in H2L and formation of unpredicted complexes 1 and 2. The crystal structure and parameters of complex 1 are similar to the previous report of our group in which the ligand undergoes similar hydrolytic cleavage [25]. Hence, here we discuss only the crystal structure of complex 2 in detail (Figs. 1, 2; Table 1). Similar to complex 1, complex 2 also displays an octahedral geometry. The coordination sites are filled up by a C=S sulfur atom, a carbonyl group, two chlorine atom and two triphenylarsine trans to each other. The ruthenium atom is in a distorted octahedral environment with trans angles of [As(2)–Ru(1)–As(1)] 176.61(4)° and [Cl(1)–Ru(1)–S(1)] 168.64(8)°. The other two axial sites are occupied by a carbonyl group and one chlorine atom with Ru(1)–C(1) and Ru(1)–Cl(1) distance of 1.803(10) and 2.573(2) Å. The carbonyl group trans to the coordinated Cl(2) atom [Cl(2)–Ru(1)–C(1)] with an angle of 176.3(3)°. This is due to strong Ru(II) → CO back donation as showed by the short Ru(1) − C(1) [1.803(10) Å] bond and low CO stretching frequency (1952/cm), which prefers σ or π weak donor groups occupying the site opposite to CO to favor the d–π back bonding donation. The second chloride ion attached to the ruthenium ion can be furnished by chloroform solvent [25]. The metal ligand bond distances (Table 2) in the complex 2 agree well with previous reports containing similar coordination environment [26]. Moreover, the complex 2 exhibited weak hydrogen bonding interactions between Cl and NH atom.
DNA interaction studies
Fluorescence emissive titration
Coordination compound and DNA interaction is one of the criterions for comprehending the molecular basis of therapeutic activity or advanced clinical trials. Generally, molecules/complexes bind non-covalently (intercalation, groove-binding and external electrostatic mode) with DNA. The metal ion, ligands and geometry of the complexes affect the binding efficiency [27]. As a basic testing method, emissive titration study is unanimously employed method to find out the binding mode of metal complexes with DNA which usually involves the changes in emissive intensity and wavelength. Emissive titration experiments were done by aliquot addition of DNA to the test compound and observe the changes in emission intensity. The intercalative mode of affinity between metal complexes and DNA results in hypochromism with or without red/blue shift; on the other hand, non-intercalative/electrostatic interaction causes hyperchromism [28]. The fluorescence emission titration curves (Fig. 3) were used to study the binding mode and propensity of complex 2 with CT-DNA in Tris–HCl buffer (5 mM Tris–HCl/50 mM NaCl buffer for pH = 7.2) at 25 °C. Upon increasing concentration of CT-DNA, the emission spectral bands of complex 2 at 323 nm exhibit hypochromism of about 21.23% with very small slight red shifts of 0.5 nm. The marked decreases in the fluorescence intensity indicate the intercalative binding mode of complex 2 and DNA. The calculated binding constant (Kb) value (Table 3) is comparable to similar type of previously reported complex [25] and also supports the intercalative mode [29]. However, some more experiments require for proving the binding mode.
Fluorescence quenching study
To gain support for the type of binding of the complex with CT-DNA, the fluorescence quenching studies were performed using metal complex as a quencher. Ethidium bromide (EB) is commonly used as sensitive fluorescent and emits enhanced emissive property in the presence of DNA [30]. From the above info in mind, the quenching of EB bound DNA is calculated with the successive addition of metal complex 2. The emission band at 686 nm for EB displays hypochromism up to 15.52% with a small red shift from the initial fluorescence intensity (Fig. 4). From this evidence, it is concluded that EB is being released from EB-DNA compound due to the reduction of the corresponding ruthenium complex. The quenching constant (Ksv) value was calculated using classical Stern–Volmer equation according to the previous literature [31], which was listed in Table 3. In addition, the following equation is used to calculate the apparent DNA-binding constant (Kapp) values:
where KEB = 1.0 × 10−7/M is the DNA-binding constant of EB, [EB] is the concentration of EB (7.5 μM) and [M50%] is the concentration of the compound used to obtain a 50% reduction in fluorescence intensity of DNA pretreated with EB. The Ksv and Kapp values are found to be comparable with our previous reported ruthenium(II) complex [25]. The results support the intercalative binding mode of complex 2 with DNA and the results are consistent with the previous emissive titrations.
Viscosity measurements
To clarify the binding mode of the Ru(II) complex with DNA, the viscosity measurements were carried out on CT-DNA by varying the concentration of added complex. The experiment involves the measurement of the flow rate of DNA solution through a capillary viscometer. For a small molecule that binds by non-classical intercalation, causes less pronounced or no change in the viscosity of DNA [32]. On the other hand, a classical intercalation mode into DNA causes a significant increase in the viscosity of DNA solution due to increase in the overall DNA molecular length. The plot of (η/η0)1/3 vs [complex]/[DNA] gives a measure of the viscosity changes (Fig. 5). The obtained plot shows increase in the relative viscosity, which clearly indicates intercalate binding mode for complexes. Therefore, we can conclude that the complex 2 binds to CT-DNA in the intercalate regions, which is in accordance with fluorescence spectral results.
Protein binding studies
The emission studies have also been used to explore the interaction of complex 2 with BSA. BSA (1 μM) was titrated with various increasing concentrations of 2 (0–50 μM) in the wave length 290–430 nm (λex = 280 nm) and their emission spectra recorded. The fluorescence spectrum of BSA exhibits a broad band with a maximum at ~ 340 nm. The outcome of increasing concentration of 2 on the emission spectra of BSA at room temperature is shown in Fig. 6a. The value of Stern–Volmer constant (Kq) calculated from following Stern–Volmer equation [33] is used to express the amount of quenching. Quenching constant (Kq) was found from the plot of log (I0 − I)/I vs log[Q] (Fig. 6a) which is much higher than the molecular fluorescence diffusing constant as shown in Table 4. The result reveals that the fluorescence quenching mechanism of the compound-BSA system will not be dynamic quenching. It may be static quenching. However, this conclusion needs to be confirmed through other experiments. The binding constant Kb was calculated using the Scatchard equation:
where Kb is the binding constant of the complex with BSA and n is the number of binding sites. The binding constant (Kb) has been obtained as per Fig. 6a and its value is given in Table 4 along with n. The value of n indicates the availability of one binding site in BSA for complex 2. The higher values of Kq and Kb signify a strong interaction between the BSA proteins. The quenching parameters for complex 2 are comparable to similar type of previously reported complex [25]. Furthermore, the static quenching fluorescence mechanism of the compound is proved by UV–Vis absorption spectral studies. Generally, quenching happens either by dynamic or static ways. When the fluorophore and the quencher come into contact during the transient existence of the excited state, dynamic quenching occurs [34]. On the other hand, static quenching represents to the formation of fluorophore–quencher compound in the ground state. UV–Vis absorption spectra of BSA have been measured in the absence and presence of 2. Figure 6b shows that addition of 2 to a fixed concentration of BSA leads to an increase in the absorbance of the BSA absorption peak at 280 nm with a small blue shift for free BSA, indicating that the complex 2 interact with BSA by the static quenching which could be attributed to the formation of the ground state complex as reported earlier [35].
In vitro cytotoxicity assay
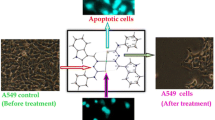
The ruthenium complex was evaluated for its cytotoxicity vs a pair of human tumor cell lines such as human cervical cancer (HeLa), human hepatocellular carcinoma (HepG2), and normal Vero cells by means of a colorimetric assay (MTT assay) that measures mitochondrial enzyme succinate dehydrogenase activity as an indication of cell viability. Cis-platin was used as the reference compound to evaluate the cytotoxic activity. The results were analyzed by cell viability curves and expressed with IC50 values of complex 2 in Table 5. The amount of cell proliferation significantly decreases in various concentration ranges (10–100 µg/cm3) on supplementation with complex, as observed within 24 h of incubation with respective of above cell lines.
From IC50 values, the complex demonstrates notable activity in human cervical cancer (HeLa), human hepatocellular carcinoma (HepG2) cell lines. However, the complex shows marginally less activity than cis-platin and comparable activity with previously reported ruthenium complexes [10, 23, 25, 26]. It is noteworthy that the complex 2 is less toxic towards the normal Vero cell line as is evident from the IC50 values.
Enzyme mimicking studies
Catecholase activity
3,5-Di-tert-butylcatachol (3,5-DTBC) was used as substrate to study the catecholase mimic activity of the complex 2. Due to the presence of two bulky tert-butyl substituents, the substrate shows a low quinine–catechol reduction potential and hence effortless oxidation occurs to the corresponding o-quinone (Scheme 2).

The stable oxidation product 3,5-di-tert-butylquinone (3,5-DTBQ) displays a maximum emission at 390 nm. Before proceeding to a thorough kinetic study, the complex 2 (1 × 10−4 M) were examined by addition of complex 2 to a 100 equivalents DMF solution of 3,5-DTBC (1 × 10−2 M) under oxygen atmosphere and the progress of the reaction was monitored for every 15 min interval up to 2 h. Owing to addition of the catecholic substrate, a new band (Fig. 7) appeared at 434 nm and its intensity steadily increased may be due to ligand to metal charge transfer band of the oxidized product 3,5-di-tert-butylbenzoquinone (3,5-DTBQ). Therefore, the experiment obviously demonstrates that the oxidation of 3,5-DTBC to 3,5-DTBQ was catalyzed by 2, as it is well known that 3,5-DTBQ displays a maximum at λemis = 434 nm in DMF. The catalytic performance exhibits saturation kinetics based on the Michaelis–Menten model appeared to be suitable under excess substrate conditions. Plots of ki vs [3,5-DTBC] provided non-linear curve of decreasing slope (Fig. 7) which are best designated kinetic equation by our earlier report [25]. The Michaelis–Menten constant (Km) and maximum initial rate (Vmax) were determined by linearization using Lineweaver–Burk plots (Fig. 7). The turnover number values (Kcat) were obtained by dividing the Vmax values by the concentration of the complex and all these parameters are listed in Table 6. The results unambiguously demonstrate that the complex 2 is active towards the oxidation of 3,5-DTBC. When compared to similar reported complex, the activity of the present complex is slightly lower although it possesses appreciable catecholase mimic activity [25]. From all the data, it is concluded that the catalytic oxidation follows the mechanism suggested by Chyn and Urbach [36].
Phosphatase activity
Based on the promising catecholase activity result, the complex was further investigated for its phosphatase activity. Phosphate esters play several important functions in biological systems such as information storage (DNA/RNA), cellular signaling (cAMP), and energy transduction (ATP) [11]. The phosphodiester bonds in these molecules are extremely resistant towards hydrolysis because of the repulsion between the negatively charged backbone and potential nucleophiles. Since the compound 4-nitrophenyl phosphate disodium salt hexahydrate (4-NPP) ester is negatively charged and under neutral conditions they are very resistant to hydrolysis, was selected as substrate. The 4-NPP hydrolysis was monitored using complex 2 as catalyst by emissive titrations (Fig. 8) on the time evolution of 4-NP in DMF at λemis = 486 nm (λex = 435 nm) over 15 min break up to 2 h with 40 equivalents of the substrate (Scheme 3).

The dependence of the initial rate on the concentration of the substrate was monitored at the respective wavelength by emission spectroscopy, which corresponds to the increase in 4-nitrophenolate (4-NP) concentration (Fig. 8). The rate vs concentration of substrate data were analyzed based on Michaelis–Menten approach of enzymatic kinetics to get the Lineweaver–Burk (double reciprocal) plot as well as the values of variety of kinetic parameters Vmax, Km and Kcat. Plot of ki vs [4-NPP] gave non-linear curve of decreasing slope which is best designated by the first-order kinetic equation.
The turnover number value (Kcat) was obtained by dividing the Vmax value by the concentration of the complex and all the parameters are listed in Table 6. The results show that the complex 2 owns superior activity and effectively catalyze the hydrolysis with appreciable turnover rate [25]. Moreover, the kinetic data reveal that the hydrolysis of 4-NPP followed the mechanism described earlier [25].
Conclusions
In conclusion, the ruthenium complexes 1 and 2 bearing thiourea ligand have been synthesized and characterized by analytical, spectral, and single crystal X-ray crystallographic techniques. The characteristic results reveal the C=N bond cleavage in ligands and monodentate coordination via C=S group with the ruthenium metal ion. The results of single crystal XRD analysis of complexes confirm ligand cleavage, monodentate coordination mode and octahedral geometry. The binding properties of the ruthenium complex with CT-DNA were evaluated by emissive titrations, fluorescence quenching, viscosity measurements, and the results show that the complex interacts with CT-DNA through intercalative mode. Furthermore, the complex BSA protein interactions were examined by UV–Vis and emissive titrations reveal that complex has shown high binding affinity and acts as static quencher. The in vitro cytotoxic study demonstrates the substantial cytotoxicity of the complex. Moreover, the complex was effectively used for aerial oxidation of catechol and phosphate hydrolysis. On the basis of Michaelis–Menton approach of enzyme kinetics, the complex shows good catalytic efficiency in catechol-oxidase functional mimic in the conversion of 3,5-DTBC to 3,5-DTBQ and ester hydrolysis functional mimic in the hydrolysis of 4-NPP to 4-NP in aerobic conditions.
Experimental
All chemicals were procured commercially and used as received. Solvents were dried and distilled following the literature procedures [37] and reactions carried out under room temperature. Microanalyses were performed using an apparatus designed by Technico Instuments, India. Vario EL III CHNS analyzer was used for elemental analysis measurement. Bruker FT IR spectrophotometer equipped with an ATR sampling accessory was employed for recording IR spectra in the range 4000–450/cm. UV–Vis spectra were recorded on Shimadzu UV-1650 PC spectrophotometer over 800–200 nm range. 1H, 13C, 31P NMR spectra were performed on a Bruker Ultra Shield using DMSO-d6 and CDCl3 as solvent. The chemical shifts are given in ppm relative to SiMe4 and orthophosphoric acid. ESI-MS were recorded on a Q-TOF micro™ mass spectrometer operating in ESI mode. Buffer solutions were prepared using doubly distilled water. Stock solution of complex (1 mM in DMSO) was stored at 4 °C and used whenever required. Data were expressed as the mean ± the standard deviation from three independent experiments. The ruthenium precursors [RuHCl(CO)(EPh3)3] (E = As or P) and the ligand (N-(N,N-diethylaminothiocarbonyl)benzimidoylchloride-2-aminoacetophenone-N-methylthiosemicarbazone) (H2L) were prepared according to literature methods [18, 38].
Synthesis of complexes 1 and 2
The metal complexes were prepared as per reported procedure with minor alteration [39]. [RuHCl(CO)(EPh3)3] (E = P or As) (1 mmol) and the ligand H2L (1 mmol) were dissolved in 10 cm3 of methanol and chloroform solvent mixture (1:1 ratio). The solution mixture was mildly heated under reflux for 12 h. The progress of the reaction was monitored by thin layer chromatography (TLC) using 9:1 mixture of petroleum ether–ethyl acetate mixture as mobile phase. After completion of the reaction, the resulting solution was filtered and the filtrate was left untroubled for slow evaporation of the solvent. After 5 days, brown colored crystals suitable for X-ray diffraction were obtained.
Carbonyldichlorido(1,1-diethylthiourea)-bis(triphenylphosphine)ruthenium(II) (1)
The analytical and spectral data are identical to the previously reported complex [25].
Carbonyldichlorido(1,1-diethylthiourea)bis(triphenylarsine)-ruthenium(II) (2, C42H42As2Cl2N2O2RuS)
Yield: 54%; color: brown; IR (KBr): = 3380 (NH2), 1939 (C≡O), 829 (C=S)/cm; 1H NMR (300.13 MHz, CDCl3): δ = 8.21 (s, NH2), 7.21–7.80 (m, aromatic), 3.11 (q, J = 7.0 Hz, 2H, CH2), 3.02 (q, J = 7.0 Hz, 2H, CH2), 1.40 (t, J = 7.0 Hz, 3H, CH3), 1.37 (t, J = 7.0 Hz, 3H, CH3) ppm; 13C NMR (300.13 MHz, CDCl3): δ = 202.12 (C≡O), 172.14 (C=S), 126.64 (Ar C), 119.24 (Ar C), 118.71 (Ar C), 112.41 (Ar C), 46.95 (CH2), 45.92 (CH2), 14.32 (CH3), 13.94 (CH3) ppm; UV–Vis (CHCl3): λmax (ε) = 238 (44,200), 266 (27,000), 339 (12,540), 443(5240) nm (dm3/mol/cm); ESI-MS: m/z calcd. 944.68, found 909.22 ([M−Cl]+).
X-ray structure determination
Crystals of complexes suitable for single crystal XRD were grown from slow evaporation of methanol–chloroform solvent mixture (CCDC reference number 1584536). Suitable single crystal of compounds with accurate dimensions was mounted on a glass fiber containing epoxy cement. Crystal data were collected on Gemini Ultra diffractometer. Structure solutions and refinements of the compounds were done using the programs SHELXS-14 [40]. Refinement and all further calculations were carried out using SHELXL. All non-solvent heavy atoms were refined anisotropically. All non-solvent hydrogen atoms were idealized using the standard SHELXL idealization methods [41]. When the non-hydrogen atoms will be refined by anisotropically, using weighted full-matrix least squares on F2. Atomic scattering factors were incorporated in the computer programs.
DNA-binding studies
The binding interaction experiments of complex with CT-DNA in double distilled water containing 95% Tris/NaCl buffer (5 mM Tris–HCl/50 mM NaCl buffer, pH 7.2) were carried out using a Fluoromax spectrofluorometer with a rectangular quartz cuvette of 1 cm path length. Stock solutions of CT-DNA used in the studies not ever exceeded 5% DMSO (v/v) in the final volume. A stock solution of CT-DNA was stored at 277 K and used after no more than 4 days. Tris–HCl buffer solution was used to base line correction. The excitation wavelength was fixed by the emission range and attuned before measurements. The emissive titration experiments were performed using a fixed concentration (25 µM) of test compounds to which increments of the (0–10 µM) DNA stock solution were added. The emission intensities were recorded for complexes in the range of 300–600 nm. Titrations were manually done by a micropipette for the addition of CT-DNA and reference solution to eliminate the absorbance of CT-DNA itself. Further support for the relative binding property of complexes binding to DNA via intercalation is given through emission quenching experiments. DNA was pretreated with ethidium bromide (EB) for 30 min. Then the test solutions were added to the mixture of EB-DNA, and the change in the fluorescence intensity was measured. The excitation and the emission wavelength were 510 nm and 604 nm, respectively. EB was non-emissive in buffer solution (pH 7.2) due to fluorescence quenching of the free EB by solvent. In the fluorescence quenching spectra, the reduction in emission intensity measured the binding mode of complexes to CT-DNA.
Viscosity experiments were carried out using an Ubbelodhe viscometer at a constant temperature (30.0 ± 0.1 °C) in a thermostat and the relative parameters were calculated from the relation η = (t − to)/to, where t is the observed flow time of DNA-containing solution and to is the flow time of Tris–HCl/NaCl buffer alone. Data were presented as (η/η0)1/3 vs binding ratio (r = [Compounds]/[DNA] = 0.0–0.1), where η is the viscosity of CT-DNA in the presence of the compound, and η0 is the viscosity of CT-DNA alone.
Protein binding studies
The excitation wavelength of BSA at 280 nm and the emission at 345 nm were monitored for the protein binding studies using fluorescence spectra recorded with synthesized compounds. The excitation wavelength of BSA at 280 nm and the quenching of the emission intensity of tryptophan residues of BSA at 346 nm were monitored using the complexes as quenchers with increasing concentrations. The excitation and emission slit widths (each 5 nm) remained constant for all the experiments. A scan rate of 200 nm/min was used. Samples were carefully degassed using pure nitrogen gas for 15 min. Stock solution of BSA was prepared in 50 µM phosphate buffer (pH 7.2) and stored in the dark at 4 °C for further use. Furthermore, the type of quenching mechanism of complexes was determined from the UV–Vis absorption spectra in the range of 200–600 nm.
In vitro anticancer activity: 3-[4,5-dimethylthiazol-2-yl]2,5-diphenyltetrazolium bromide (MTT) assay
The human cervical cancer (HeLa), human hepatocellular carcinoma (HepG2) were obtained from National Centre for Cell Science (NCCS), Pune and grown in Eagles Minimum Essential Medium containing 10% fetal bovine serum (FBS). The cells were maintained at 37 °C, 95% air and 100% relative humidified atmosphere. The cytotoxicity of the investigated ruthenium(II) complex in comparison to cisplatin was determined using the MTT assay. The MTT colorimetric assay is based on the measurement of mitochondrial enzyme succinate dehydrogenase activity, as an indication of cell viability. MTT is a yellow water-soluble tetrazolium salt. A mitochondrial enzyme in living cells, succinate dehydrogenase, cleaves the tetrazolium ring, converting the MTT to an insoluble purple formazan. Therefore, the amount of formazan produced is directly proportional to the number of viable cells. After 48 h of incubation, 15 µM of MTT (5 mg/cm3) in phosphate buffered saline (PBS) was added to each well and incubated at 37 °C for 4 h. The medium with MTT was then flicked off and the formed formazan crystals were solubilized in 100 µM of DMSO and then measured the absorbance at 570 nm using microplate reader. One hundred microliters per well of cell suspension were seeded into 96-well plates at plating density of 10,000 cells/well and incubated to allow for cell attachment at 37 °C, 5% CO2, 95% air, and 100% relative humidity. After 24 h the cells were treated with serial concentrations of the test samples. They were initially dissolved in DMSO and an aliquot of the sample solution was diluted to twice the desired final maximum test concentration with serum free medium. Additional four serial dilutions were made to provide a total of five sample concentrations. Aliquots of 100 mm3 of these different sample dilutions were added to the appropriate wells already containing 100 mm3 of medium, resulting in the required final sample concentrations. Following sample addition, the plates were incubated for an additional 48 h at 37 °C, 5% CO2, 95% air, and 100% relative humidity. The medium containing without samples was served as control and triplicate was maintained for all concentrations. Non-linear regression graph was plotted between % cell inhibition and log concentration and IC50 was determined using Graph Pad Prism software.
Enzyme kinetic studies
The kinetics for the oxidation of catalytic oxidation of 3,5-ditertbutylcatechol (3,5-DTBC) and the hydrolysis of 4-nitrophenyl phosphate (4-NPP) were monitored by fluorescence spectra under pseudo first-order kinetic reaction. 100 equivalents of 3,5-DTBC and 40 equivalents of 4-NPP in DMF (10−3 M) were added to 10−4 M solutions of ruthenium(II) complex under aerobic conditions. Emissive intensity of the resultant reaction mixture was plotted with respect to wavelength at a regular interval of 15 min using a fluorescence spectrophotometer in the range 410–700 nm. The dependence of the rate on substrate concentration and diverse kinetic parameters were obtained by treatment of complexes with 3,5-DTBC and 4-NPP monitoring the increase in emission intensity at 435 and 485 nm, respectively (the peak corresponding to the band maxima), as a function of time.
References
Alderden RA, Hall MD, Hambley TW (2006) J Chem Educ 83:728
Kelland L (2007) Nat Rev Cancer 7:573
Breydo L, Uversky VN (2011) Metallomics 3:1163
Barry NPE, Sadler PJ (2013) Chem Commun 49:5106
Li VS, Choi D, Wang Z, Jimenez LS, Tang MS, Kohn H (1996) J Am Chem Soc 18:2326
Zuber G, Quada JC, Hecht SM (1998) J Am Chem Soc 120:9368
Liu JG, Ye BH, Li H, Zhen QX, Ji LN, Fu YH (1999) J Inorg Biochem 76:265
Santini C, Pellei M, Gandin V, Porchia M, Tisato F, Marzano C (2014) Chem Rev 114:815
Suess-Fink G (2010) Dalton Trans 39:1673
Vijayan P, Viswanathamurthi P, Sugumar P, Ponnuswamy MN, Balakumaran MD, Kalaichelvan PT, Velmurugan K, Nandhakumar R, Butcher RJ (2015) Inorg Chem Front 2:620
Sanyal R, Guha A, Ghosh T, Mondal TK, Zangrando E, Das D (2014) Inorg Chem 53:85
Mukherjee J, Mukherjee R (2002) Inorg Chim Acta 337:429
Neves A, Rossi LM, Bortoluzzi AJ, Szpoganicz B, Wiezbicki C, Schwingel E (2002) Inorg Chem 41:1788
Gomathi A, Vijayan P, Viswanathamurthi P, Suresh S, Nandhakumar R, Hashimoto T (2017) J Coord Chem 70:1645
Nguyen HH, Abram U (2009) Eur J Inorg Chem 3179
Nguyen HH, Abram U (2007) Inorg Chem 46:5310
Castillo Gomez JD, Hagenbach A, Gerling-Driessen UIM, Koksch B, Beindorff N, Brenner W, Abram U (2017) Dalton Trans 46:14602
Vijayan P, Viswanathamurthi P, Velmurugan K, Nandhakumar R, Balakumaran MD, Malcki JG (2015) RSC Adv 5:103321
Barandov A, Abram U (2009) Inorg Chem 48:8072
Manikandan R, Anitha P, Prakash G, Vijayan P, Viswanathamurthi P (2014) Polyhedron 81:619
Nguyen HH, Pham CT, Abram U (2015) Inorg Chem 54:5949
Maia PIS, Nguyen HH, Hagenbach A, Bergemann S, Gust R, Deflon VM, Abram U (2013) Dalton Trans 42:5111
Sathiya Kamatchi T, Chitrapriya N, Lee H, Fronczek CF, Fronczek FR, Natarajan K (2012) Dalton Trans 41:2066
Manikandan R, Anitha P, Prakash G, Vijayan P, Viswanathamurthi P, Butcher RJ, Malecki JG (2015) J Mol Catal A Chem 398:312
Vijayan P, Viswanathamurthi P, Sugumar P, Ponnuswamy MN, Malecki JG, Velmurugan K, Nandhakumar R, Balakumaran MD, Kalaichelvan PT (2017) Appl Organometal Chem 31:e3652
Kalaivani P, Prabhakaran R, Poornima P, Dallemer F, Vijayalakshmi K, Vijaya Padma V, Natarajan K (2012) Organometallics 31:8323
Paitandi RP, Gupta RK, Singh RS, Sharma G, Koch B, Pandey (2014) Eur J Med Chem 84:17
Jaividhya P, Dhivya R, Akbarsha MA, Palaniandavar M (2012) J Inorg Biochem 114:94
Rajarajeswari C, Loganathan R, Palaniandavar M, Suresh E, Riyasdeen A, Akbarshae MA (2013) Dalton Trans 42:8347
Almes FJM, Porschke D (1993) Biochemistry 32:4246
Baron ESG, DeMeio RH, Klemperer F (1936) J Biol Chem 112:625
Liu YJ, Liang JH, Li ZZ, Yao JH, Huang HL (2011) J Organomet Chem 696:2728
Rajendiran V, Karthik R, Palaniandavar M, Evans HS, Periasamay VS, Akbarsha MA, Srinag BS, Krishnamurthy H (2007) Inorg Chem 46:8208
Yu Q, Liu Y, Zhang J, Yang F, Sun D, Liu D, Zhou Y, Liu J (2013) Metallomics 5:222
Lackowicz JR, Weber G (1973) Biochem J 12:4161
Chyn JP, Urbach FL (1991) Inorg Chim Acta 189:157
Vogel AI (1989) Text book of practical organic chemistry. Longman, London
Ogiwara Y, Miyake M, Kochi T, Kakiuchi F (2017) Organometallics 36:159
Vijayan P, Viswanathamurthi P, Silambarasan V, Velmurugan D, Velmurugan K, Nandhakumar R, Butcher RJ, Silambarasan T, Dhandapani R (2014) J Organomet Chem 768:163
SMART & SAINT (1998) Software reference manuals, version 5.0. Bruker AXS Inc, Madison
Sheldrick GM (1997) SHELXL97. Program for Crystal Structure Refinement. University of Gottingen, Gottingen
Acknowledgements
Open access funding provided by TU Wien (TUW). The authors thank UGC, India for financial assistance to Department of Chemistry, Periyar University, Salem under SAP (no. 540/20/DRS-I/2016(SAP-I)).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Vijayan, P., Vijayapritha, S., Ruba, C. et al. Ruthenium(II) carbonyl complexes containing thiourea ligand: Enhancing the biological assets through biomolecules interaction and enzyme mimetic activities. Monatsh Chem 150, 1059–1071 (2019). https://doi.org/10.1007/s00706-019-2357-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-019-2357-5