Abstract
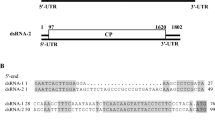
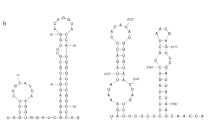
The complete nucleotide sequence of a novel mycovirus, designated as "Rhizoctonia fumigata bipartite virus 1" (RfBV1), from Rhizoctonia fumigata AG-Ba isolate C-314 Baishi was determined. The genome of RfBV1 is composed of two double-stranded RNAs (dsRNA). dsRNA-1 (2311 bp) contains one open reading frame (ORF), which codes for the putative RNA-dependent RNA polymerase (RdRp) of the virus. dsRNA-2 (1690 bp) contains one ORF, which encodes a putative protein whose function is unknown. Phylogenetic analysis indicated that the RdRp of RfBV1 clustered with several unassigned bipartite viruses belonging to the CThTV-like viruses group, but not the family Amalgaviridae or Partitiviridae. Our study suggests that RfBV1 is a novel mycovirus related to the CThTV-like viruses.


Similar content being viewed by others
References
Ogoshi A (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kuhn. Ann Rev Phytopathol 25:125–143
Castanho B, Butler EE, Shepherd RJ (1978) The association of double-stranded RNA with Rhizoctonia decline. Phytopathology 68:1515–1519
Castanho B, Butler EE (1978) Rhizoctonia decline: a degenerative disease of Rhizoctonia solani. Phytopathology 68:1505–1510
Abdoulaye AH, Foda MF, Kotta-Loizou I (2019) Viruses infecting the plant pathogenic fungus Rhizoctonia solani. Viruses. https://doi.org/10.3390/v11121113
Zheng L, Liu H, Zhang M, Cao X, Zhou E (2013) The complete genomic sequence of a novel mycovirus from Rhizoctonia solani AG-1 IA strain B275. Arch Virol 158:1609–1612
Zheng L, Shu C, Zhang M, Yang M, Zhou E (2019) Molecular Characterization of a novel Endornavirus conferring hypovirulence in rice sheath blight fungus Rhizoctonia solani AG-1 IA strain GD-2. Viruses. https://doi.org/10.3390/v11020178
Zhang M, Zheng L, Liu C, Shu C, Zhou E (2018) Characterization of a novel dsRNA mycovirus isolated from strain A105 of Rhizoctonia solani AG-1 IA. Arch Virol 163:427–430
Lyu R, Zhang Y, Tang Q, Li Y, Cheng J, Fu Y, Chen T, Jiang D, Xie J (2018) Two alphapartitiviruses co-infecting a single isolate of the plant pathogenic fungus Rhizoctonia solani. Arch Virol 163:515–520
Liu C, Zeng M, Zhang M, Shu C, Zhou E (2018) Complete nucleotide sequence of a partitivirus from Rhizoctonia solani AG-1 IA strain C24. Viruses. https://doi.org/10.3390/v10120703
Picarelli M, Forgia M, Rivas EB, Nerva L, Chiapello M, Turina M, Colariccio A (2019) Extreme diversity of mycoviruses present in isolates of Rhizoctonia solani AG 2–2 LP from Zoysia japonica from Brazil. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2019.00244
Li S, Li Y, Hu C, Han C, Zhou T, Zhao C, Wu X (2020) Full genome sequence of a new mitovirus from the phytopathogenic fungus Rhizoctonia solani. Arch Virol 165:1719–1723
Das S, Falloon RE, Stewart A, Pitman AR (2016) Novel mitoviruses in Rhizoctonia solani AG-3PT infecting potato. Fungal Bio 120:338–350
Das S, Das S (2019) First report of a novel alphapartitivirus in the basidiomycete Rhizoctonia oryzae-sativae. Arch Virol 164:889–892
Chen Y, Gai XT, Chen RX, Li CX, Zhao GK, Xia ZY, Zou CM, Zhong J (2019) Characterization of three novel betapartitiviruses co-infecting the phytopathogenic fungus Rhizoctonia solani. Virus Res. https://doi.org/10.1016/j.virusres.2019.197649
Chen Y, Su JE, Qin XY, Fan ZY, Zhang XH, Yu Q, Xia ZY, Zou CM, Zhao GK, Lin ZL (2020) A novel putative betapartitivirus isolated from the plant-pathogenic fungus Rhizoctonia solani. Arch Virol 165:1697–1701
Charlton ND, Carbone I, Tavantzis SM, Cubeta MA (2008) Phylogenetic relatedness of the M2 double-stranded RNA in Rhizoctonia fungi. Mycologia 100:555–564
Li W, Zhang T, Sun H, Deng Y, Zhang A, Chen H, Wang K (2014) Complete genome sequence of a novel endornavirus in the wheat sharp eyespot pathogen Rhizoctonia cerealis. Arch Virol 159:1213–1216
Li Y, Xu P, Zhang L, Xia Z, Qin X, Yang G, Mo X (2015) Molecular characterization of a novel mycovirus from Rhizoctonia fumigata AG-Ba isolate C-314 Baishi. Arch Virol 160:2371–2374
Li Y (2015) The establishment of a new anastomosis group AG-V and research of RNA viruses from Rhizoctonia spp., Dissertation, Yunnan Agricultural University
Morris T, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Froussard P (1992) A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res 20:2900
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Liu C, Li M, Redda ET, Mei J, Zhang J, Elena SF, Wu B, Jiang X (2019) Complete nucleotide sequence of a novel mycovirus from Trichoderma harzianum in China. Arch Virol 164:1213–1216
Liu H, Wang H, Zhou Q (2021) A novel mycovirus isolated from the plant-pathogenic fungus Botryosphaeria dothidea. Arch Virol 166:1267–1272
Marquez LM, Redman RS, Rodriguez RJ, Roossinck MJ (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513–515
Sutela S, Vainio EJ (2020) Virus population structure in the ectomycorrhizal fungi Lactarius rufus and L. tabidus at two forest sites in Southern Finland. Virus Res. https://doi.org/10.1016/j.virusres.2020.197993
Yu J, Kwon SJ, Lee KM, Son M, Kim KH (2009) Complete nucleotide sequence of double-stranded RNA viruses from Fusarium graminearum strain DK3. Arch Virol 154:1855–1858
Wang C, Wu J, Zhu X, Chen J (2016) Complete nucleotide sequences of dsRNA2 and dsRNA7 detected in the phytopathogenic fungus Sclerotium hydrophilum and their close phylogenetic relationship to a group of unclassified viruses. Virus Genes 52:823–827
Wang L, Wang S, Yang X, Zeng H, Qiu D, Guo L (2017) The complete genome sequence of a double-stranded RNA mycovirus from Fusarium graminearum strain HN1. Arch Virol 162:2119–2124
Wang Y, Zhao H, Xue C, Xu C, Geng Y, Zang R, Guo Y, Wu H, Zhang M (2020) Complete genome sequence of a novel mycovirus isolated from the phytopathogenic fungus Corynespora cassiicola in China. Arch Virol 165:2401–2404
Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M (2016) Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res 219:22–38
Acknowledgements
The authors would like to thank Dr. A. Ogashi from Hokkaido University, Japan, for kindly providing the Rhizoctonia fumigata AG-Ba isolate C-314 Baishi used in this study. This research was supported by Kunming University (YJL16006), Yunnan Provincial Department of Science and Technology (2018FH001-032, 2018FH001-037, 2018FD080), the National Natural Science Foundation of China (31960525), and Ningbo University (KF20190107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Handling Editor: Ioly Kotta-Loizou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The nucleotide sequence data reported here are available in the DDBJ/EMBL/GenBank databases under the accession numbers KP209316 and KP209317.
Rights and permissions
About this article
Cite this article
Li, Y., Xu, P., Zhang, L. et al. Complete nucleotide sequence of a novel mycovirus infecting Rhizoctonia fumigata AG-Ba isolate C-314 Baishi. Arch Virol 167, 959–963 (2022). https://doi.org/10.1007/s00705-021-05269-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05269-7