Abstract
H5N1 highly pathogenic avian influenza viruses (HPAIVs) are a threat to both animal and public health and require specific and rapid detection for prompt disease control. We produced three neutralizing anti-hemagglutinin (HA) monoclonal antibodies (mAbs) using two clades (2.2 and 2.5) of the H5N1 HPAIV isolated in Japan. Blocking immunofluorescence tests showed that each mAb recognized different epitopes; 3B5.1 and 3B5.2 mAbs against the clade 2.5 virus showed cross-clade reactivity to all 26 strains from clades 1, 2.2, 2.3.2.1, 2.3.2.1a, b, c and 2.3.4, suggesting that the epitope(s) recognized are conserved. Conversely, the 1G5 mAb against the clade 2.2 virus showed reactivity to only clades 1, 2.3.4 and 2.5 strains. An analysis of escape mutants, and some clades of the H5N1 viruses recognized by 3B5.1 and 3B5.2 mAbs, suggested that the mAbs bind to an epitope, including amino acid residues at position 162 in the HA1 protein (R162 and K162). Unexpectedly, however, when five Eurasian-origin H5 low-pathogenic AIV (LPAIV) strains with R162 were examined (EA-nonGsGD clade) as well as two American-origin strains (Am-nonGsGD clade), the mAb recognized only EA-nonGsGD clade strains. The R162 and K162 residues in the HA1 protein were highly conserved among 36 of the 43 H5N1 clades reported, including clades 2.3.2.1a and 2.3.2.1c that are currently circulating in Asia, Africa and Europe. The amino acid residues (158-PTIKRSYNNTNQE-170) in the HA1 protein are probably an epitope responsible for the cross-clade reactivity of the mAbs, considering the epitopes reported elsewhere. The 3B5.1 and 3B5.2 mAbs may be useful for the specific detection of H5N1 HPAIVs circulating in the field.
Similar content being viewed by others
Introduction
The first outbreak of H5N1 highly pathogenic avian influenza viruses (HPAIVs) occurred in Hong Kong in 1997 in chicken populations. Eighteen human cases with 33.3% mortality were also observed at the same time [1]. Since then, these viruses have continued to cause deadly poultry outbreaks in Asia, Africa and Europe [2]. They have also caused serious diseases associated with high mortality in humans, leading to an increased awareness of the potential for a pandemic outbreak [3]. Recently, H5N1 viruses also appeared in North America [4].
Currently, HPAIVs have been classified into clades 0–9 based on phylogenetic analysis of their hemagglutinin (HA) genes [1, 5]. However, continued evolution of H5N1 viruses has led to the emergence of new phylogenetic groups, except for clades 0, 3, 4, 5, 6, 8, and 9 that have not been recently detected [5–8]. Clades 1, 2 and 7 have been phylogenetically divided into additional clades. In particular, clade 2 viruses have evolved rapidly and extensively, and the continued evolution of the virus has generated second-order (clades 2.1–2.5), third-order (2.1.1–2.1.3, 2.2.1, 2.2.2, 2.3.1–2.3.4), fourth-order (2.1.3.1–2.1.3.3, 2.2.1.1, 2.2.1.2, 2.2.2.1, 2.3.2.1 and 2.3.4.1–2.3.4.4), and fifth-order groups (2.1.3.2a, b, 2.2.1.1a, and 2.3.2.1a, b, c). Some new fifth-order clade group strains (2.3.2.1a, c) have caused outbreaks in Asia, and recently, these clade strains have been detected in Africa and Europe [9–11]. This suggests that the newly emerging H5N1 HPAIVs are circulating in many regions. However, not all strains of this H5 subtype are highly pathogenic: only a small number of H5 low pathogenic AIV (LPAIV) have mutated to the HPAIV form [12]. LPAIVs can be classified into Eurasian and American clades (EA-nonGsGD and Am-nonGsGD) that originated in Asia and America, respectively [13]. As a result, an accurate, rapid, and specific diagnostic system for HPAIV and LPAIV infection is essential for the establishment of prompt disease control.
The currently available diagnostic techniques for identifying H5N1 viruses, e.g. virus isolation followed by serological tests to determine HA and NA subtypes or reverse transcription polymerase chain reaction, have some disadvantages in terms of high cost, rapidness, necessary expertise, and the need for a biosafety level- (BSL) 3 laboratory [14]. Rapid diagnostic tests using monoclonal antibodies (mAbs) against H5N1 HA proteins (anti-H5 mAbs) to immunologically detect H5N1 viruses have been reported [14–16]. However, there have been no reports as to whether the mAbs used in these tests could detect currently circulating H5N1 clade viruses, although the tests are based on the use of broad, cross-reactive, H5-specific mAbs. Therefore, it is important to continuously make efforts to search for anti-H5 mAbs with broad reactivity against HA proteins for the development of diagnostic systems that are highly sensitive and can rapidly detect H5N1 HPAIVs, especially those H5N1 viruses that are currently circulating as well as the newly emerging H5N1 clades.
Recently, anti-H5 mouse and human mAbs were demonstrated to recognize broad cross-reactive epitopes in several clades of viruses: 1, 2.1.3.2, 2.2, and 2.3.4 [17], 0, 1, 2.2, 2.3.2.1, and 2.3.4 [18], and 0–9, except clade 7.2 [19]. Unfortunately, the newly circulating H5N1 clades 2.3.2.1a and 2.3.2.1c were not evaluated [17–19]. Recently, Xiong et al. [20] reported that broad cross-reactive anti-H5 human mAbs reacted to clade 0, 1 2.1.3, 2.2, 2.2.1, 2.3.2, 2.3.2.1a, 2.3.4, and 2.5 viruses; however, some strains of clade 2.3.2.1a were not recognized by these mAbs.
Here, we tried to produce anti-H5 mAbs specific for H5N1 HPAIVs and evaluate cross-clade reactivity using H5N1 HPAIV strains isolated in Japan and Vietnam in addition to Eurasian-H5 (clade EA-nonGsGD) and American-H5 (clade Am-nonGsGD) LPAIV strains. We also analyzed the possible antigenic sites on the HA proteins for the anti-H5 mAbs established in this study.
Materials and methods
Viruses and cells
A total of 38 strains of influenza viruses, including different clades of H5N1 HPAIVs isolated in Japan and Vietnam, H5N1 escape mutant viruses, low pathogenic avian influenza viruses (LPAIVs), and human influenza A virus was used in this study (Table 1). All viruses were propagated in the allantoic cavities of 10-day-old embryonated chicken eggs. All experiments with the viruses were conducted in a BSL-3 laboratory approved by the relevant committee at our institution.
Madin-Darby canine kidney (MDCK) cells and mouse myeloma cells (Sp2/0) were cultured as reported previously [21, 22].
Production of mAbs
Ck/Yamaguchi/7/04 (clade 2.5) and Ck/Miyazaki/K11/07 (clade 2.2) were purified as described previously [21] and inactivated with 0.1% formalin. Two female BALB/c mice (8-weeks-old) were immunized with 0.2 mL of the inactivated purified H5N1 viruses, emulsified in a squalene-based adjuvant (AddaVAX™, InvivoGen, San Diego, CA). The inoculum, at a HA titer of 1,280, was delivered by intraperitoneal injection three times at intervals of 2–3 weeks. Final immunization was performed intravenously with the same viruses in 0.1 mL of phosphate buffered saline (PBS, pH 7.4), but without the adjuvant, three days before cell fusion. Fusion of the mouse spleen cells with the SP2/0 cells was performed, and the fused cells were cultivated as previously described [22], except that an OPI media supplement (Sigma-Aldrich Japan) and an endothelial cell growth supplement (Sigma-Aldrich Japan) were applied. Antibody-producing hybridomas were first screened using an enzyme-linked immunosorbent assay (ELISA) and then anti-HA antibody-producing hybridomas were selected using a hemagglutination inhibition test (HIT), as described below. The anti-HA antibody-producing hybridomas were cloned by a single cell pick up method, and these hybridomas were injected into the peritoneal cavity of mice to produce ascitic fluids containing mAbs, as described previously [23]. The IgG fractions of these ascitic fluids were precipitated with a 50% saturated ammonium sulphate solution, followed by dialysis against PBS.
All mouse studies were conducted in compliance with the institutional rules for the care and use of laboratory animals using protocols approved by the relevant committee at the institution.
ELISA
ELISA was performed as previously described [24] with some modifications. Briefly, the inactivated purified viruses were incubated with a lysis buffer (4% Triton-X100/2M KCl) at 4 °C for 30 min, and coated onto wells of ELISA microplates (Thermo Fisher Scientific, K.K., Yokohama, Japan) at 4 °C overnight. The plates were blocked with 10% newborn calf serum in PBS at 37 °C for 2 h. Culture supernatants of the hybridomas were then added to each well and incubated at 37 °C for 1 h. Peroxidase-conjugated goat anti-mouse polyvalent Igs (IgA, IgG, and IgM) (Sigma-Aldrich, Japan) were then added and incubated at 37 °C for 1 h. A TMB peroxidase substrate reagent (BD Bioscience Pharmingen, San Diego, CA) was added to each well and the optical density (OD) of the samples was measured at 450 nm.
Hemagglutination inhibition test (HIT)
HITs were performed according to the Manual on Animal Influenza Diagnosis and Surveillance [25] using 0.5% chicken red blood cells. Briefly, serial 2-fold dilutions of the receptor destroying enzyme-treated-ascitic fluids containing mAbs were mixed with 4 HA units/25 µL of the viruses, and then incubated at room temperature for 30 min. The chicken red blood cells were then added to the mixtures and incubated for 30 min. The HI titer was examined as a reciprocal of the highest dilution of mAb that completely inhibited hemagglutination.
Indirect fluorescence antibody test (IFAT) and blocking IFAT
IFATs were used to determine the mAbs isotypes. Briefly, MDCK cells were inoculated with the Ck/Yamaguchi/7/04 or the Ck/Miyazaki/K11/07 and incubated for 18 h. The infected cells were fixed with acetone for 10 min and incubated with mAbs (1:100) for 1 h. The infected cells were then incubated with fluorescein isothiocyanate- or Rhodamine-conjugated anti-mouse IgG subclasses (IgG1, IgG2a, or IgG2b), (Rockland Immunochemicals, Gilbertsvile, PA).
In blocking IFATs, infected cells were incubated with the 3 mAbs (3B5.1, 3B5.2, or 1G5) produced in this study for 30 min at 37 °C. Subsequently, the 3B5.1 mAb labeled with Alexa Fluor® 488 using a Fluorescein Labeling kit (Dojindo Molecular Technologies, Inc., Tokyo, Japan) or the 3B5.2 and 1G5 mAbs labeled with Alexa Fluor® 594 using a Zenon® Mouse IgG Labeling kit (Thermos Fisher Scientific, K.K.) were incubated for 30 min. The fluorescent signal was observed under a fluorescence microscope (Biorvo BZ-9000, Keyence, Japan).
Western blotting (WB) analysis
The inactivated purified viruses were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis with or without reduction, and transferred to polyvinylidene difluoride membranes using a semi-dry blotting apparatus. The membranes were blocked with 3% bovine serum albumin in PBS and incubated with ascitic fluids containing mAbs. Peroxidase-conjugated goat anti-mouse polyvalent Igs (IgA, IgG, and IgM) (Sigma-Aldrich Japan) were used as secondary antibodies. The membrane was developed with enhanced chemiluminescence WB detection reagents (GE Healthcare UK Ltd., Buckinghamshire, UK) and photographed using an LAS-3000 image analyzer (Fujifilm, Tokyo, Japan).
Virus neutralization test (VNT)
VNTs with a constant mAb concentration and diluted virus were performed. Briefly, serial 10-fold stepwise dilutions of the virus were mixed with mAb (1:10) or virus growth medium (VGM) [21] and incubated for 2 h at 37 °C. Subsequently, the mixtures were added to the MDCK cells grown in 96-well microplates and incubated for 2 h. After four days, the 50% tissue culture infectious dose (TCID50) of the mixture was determined [26]. The neutralizing index was calculated by subtracting the log TCID50 of the mAb-virus mixture from that of the virus control. mAbs with a neutralizing index higher than 0.7 were regarded to have neutralizing activity.
Selection and sequencing of escape mutants
Serial dilutions of the virus were incubated with an excess of mAb for 1 h, and the mixtures were inoculated onto MDCK cells and incubated for 3 d at 37 °C. Escape mutants were cloned by limiting-dilution in embryonated chicken eggs. Viral RNAs were extracted from the mutant-infected allantoic fluids using ISOGEN II (NIPPON GENE, Tokyo, Japan). The construction and purification of cDNA libraries, which were used for next-generation sequencing, were conducted according to a previous report [27]. The sequencing was carried out on a MiSeq bench-top sequencer (Illumina, San Diego, CA) to generate 51-bp single-end reads. The FASTQ-formatted sequence data was generated using the MiSeq Reporter program (Illumina). The contiguous sequences were assembled from the short sequence reads using CLC Genomics Workbench version 6.5.1 (CLC bio, Aarhus, Denmark). The consensus sequence was determined using BLAST.
Analysis of epitopes recognized by mAbs
The HA1 amino acid sequences (positions 1-322, corresponding to canonical H5 numbering) of the parental strains (Ck/Yamaguchi/7/04 and Ck/Miyazaki/K11/07) and their escape mutants (3B5.1, 3B5.2, and 1G5mts) were aligned and the mutation sites in the HA1 protein were further analyzed.
The 3D-structural model of the HA1 protein of the Ck/Yamaguchi/7/04 strain was constructed by using the Phyre2 web server [28], based on homology modeling. The HA1 crystal structure of the A/Vietnam/1194/04 strain was downloaded from the protein data bank (PDB) (PDB ID: 2IBXA) and used as the template for this construction.
The sixteen HA1 amino acid sequences of the different clades of H5N1 HPAIVs, escape mutants, and H5 subtype LPAIVs used in this study were aligned to predict a cross-clade epitope recognized by the anti-H5 mAbs. Subsequently, the predicted cross-clade epitope was analyzed and mapped to the HA1 structural model of the Ck/Yamaguchi/7/04 strain using PyMOL [29]. A predicted cross-clade epitope was further analyzed to find conserved amino acid residues in the HA1 protein, among the 5,366 HA amino acid sequences from H5N1 viruses clades 0–9 obtained from GenBank and the Influenza Research Database.
Results
Production and characterization of mAbs
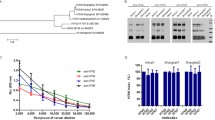
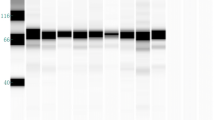
Three hybridomas (anti-H5 mAbs; 3B5.1, 3B5.2, and 1G5) with both HI and neutralizing activities were established (Fig. 1A). The 3B5.1 and 3B5.2 mAbs to Ck/Yamaguchi/7/04 (clade 2.5) showed higher HI titers and neutralizing indexes than the 1G5 mAb to Ck/Miyazaki/K11/07 (clade 2.2).
Characterization of anti-H5 mAbs to H5N1 HPAIV. (A). The general properties of the anti-H5 mAbs (3B5.1, 3B5.2 and 1G5), including HIT, VNT and antibody isotypes are shown. *The HIT and VNT assays were performed using the homologous viral strain. Identification of anti-H5 mAbs by Western blot analysis using the inactivated purified Ck/Yamaguchi/7/04 (clade 2.5) and Ck/Miyazaki/K11/07 (clade 2.2) strains as antigens under reducing and non-reducing conditions (B). One anti-H5 mAb (3B5.1) had reactivity to the HA1 protein (50 KDa) under reducing conditions and to HA proteins (75 KDa) under non-reducing condition (Ck/Yamaguchi/7/04). Two anti-H5 mAbs (3B5.2 and 1G5) had reactivity to HA proteins (75 KDa) (Ck/Yamaguchi/7/04 and Ck/Miyazaki/K11/07 strains). 1203/4: anti-H5 mAb to A/Vietnam/1203/4; M: molecular marker; +: reducing condition; -: non-reducing condition; Bl: Blank; Ya: inactivated purified Ck/Yamaguchi/7/04 antigen; Mi: inactivated purified Ck/Miyazaki/K11/07 antigen
WB analysis showed that the 3B5.2 and 1G5 mAbs reacted to a 75 kDa protein band corresponding to HA0 only under non-reducing conditions, indicating that these mAbs recognized a discontinuous (conformational) epitope on the HA proteins. However, the 3B5.1 mAb recognized both 75 and 50 kDa proteins of HA (HA0 and HA1) under non-reducing and reducing conditions, respectively (Fig. 1B). This suggested that the 3B5.1 mAb specifically recognized linear and conformational epitopes on the HA proteins.
Reactivity of the mAbs with heterologous H5 subtype influenza viruses
The 3B5.1 and 3B5.2 mAbs against Ck/Yamaguchi/7/04 (clade 2.5) had reactivity to all 26 heterologous strains examined from clades 1, 2.2, 2.3.2.1, 2.3.2.1a, 2.3.2.1b, 2.3.2.1c, and 2.3.4 of H5N1 HPAIV, isolated in Japan and Vietnam. The HI titers of the 3B5.1 mAb to these heterologous H5N1 viruses ranged from 160–640, and those of the 3B5.2 mAb ranged from 10–640 (Table 2). In contrast, the 1G5 mAb against Ck/Miyazaki/K11/07 (clade 2.2) recognized only clade 1, 2.3.4 and 2.5 viruses with the HI titers ranging from 20–320. However, only one of the five strains examined in clade 2.3.4 reacted with this mAb (titer of 1:20).
The 3B5.1, 3B5.2, and 1G5 mAbs also recognized the H5N1, H5N2, and H5N3 LPAIV strains belonging to the EA-nonGsGD clade with the HI titers ranging from 80 to 1,280, except for the Ck/Ibaraki/1/05 (H5N2) and Whis. sw./Shimane/499/83 (H5N3) strains belonging to the Am-nonGsGD clade (Table 3).
Blocking IFAT
Blocking IFAT was performed to verify whether the three anti-H5 mAbs recognized different antigenic sites on the HA1 protein. Binding of the fluorescein-conjugated 3B5.1, 2B5.2, and 1G5 mAbs was not blocked by any competitor mAbs, except for the homologous mAbs (Fig. 2), indicating that each of the mAbs recognized different antigenic sites.
Blocking immunofluorescence antibody tests. Blocking tests were conducted using anti-H5 mAbs (3B5.1, 3B5.2, and 1G5) as competitors. The mAbs were directly labeled with green or red flurophores. The 3B5.1 mAb was conjugated with Alexa Fluor® 488; 3B5.2 with Alexa Fluor® 594; and 1G5 with Alexa Fluor® 594
Analysis of the escape mutants and epitopes recognized by the mAbs
Three escape mutants to the anti-H5 mAbs (3B5.1 mt, 3B5.2 mt, and 1G5 mt) were generated and sequenced to identify the mutated amino acid positions in H5 HA1 that were potentially associated with the mAbs’ binding sites (Fig. 3A). The 3B5.1 mt carried point mutations at amino acid positions 56 (S56N), and 162 (R162I) (according to mature H5 numbering), whereas the 3B5.2 mt had two mutation sites at positions 162 (R162G) and 184 (A184G) in the HA1 protein. The 1G5mt had only a single point mutation at position 139 (G139R).
Characterization of the escape mutants (A), *selected for by use of mAbs against Ck/Yamaguchi/7/04, **selected for by use of mAbs against Ck/Miyazaki/K11/07. The mutated HA amino acid residues and antigenic sites of: the 3B5.1 mt and 3B5.2 mt displayed on the HA1 structural protein model of Ck/Yamaguchi/7/04 (B); and the 1G5 mt on the Ck/Miyazaki/K11/07 strain model (C). Two HA1 amino acid domains from the Ck/Yamaguchi/7/04 strain are located adjacent to the amino acid at position 162 (D). The HA1 amino acid position 162 is labeled in blue. The amino acids at positions 115-QIIPKSSWSDHEAS-128 are labeled magenta, while the amino acids at 158-PTIKRSYNNTNQE-170 are labeled yellow (color figure online)
Subsequently, these mutated amino acid positions were mapped to the HA1 crystal structure (2IBXA). The result showed that the three mutated amino acid residues (positions 139, 162 and 184) were located on the HA1 protein (Fig. 3B and C). The mutated amino acid at position 139 was located in antigenic site A in H3 [37] and Ca in H1 HA [38], whereas the mutated amino acid at position 184 was located in antigenic site B in H3 [37] and Sb in H1 HA [38]. The mutated amino acid at position 162 was located in antigenic site Sa in H1 HA [38]. However, the mutated amino acid at position 56 was not located in any recognized antigenic sites of H1 and H3 HA (Fig. 3B). The HA amino acid residue at position 184 (E184) in clades 2.3.2.1, 2.3.2.1a and 2.3.2.1c used in this study (data not shown) was different from that of the parental strain (G184), but these clade strains were recognized by the 3B5.1 and 3B5.2 mAbs (Table 2). Altogether, these results suggested that the HA amino acid residue at position 162 is a major epitope recognized by the 3B5.1 and 3B.2 mAbs. The 1G5 mAb recognizes the amino acid residue at position 139 in the HA protein.
In this study, a 3D-structural model of the Ck/Yamaguchi/7/04 strain was analyzed to identify the binding site of 3B5.1 and 3B5.2 on the HA1 protein. Two different amino acid residue domains were found to be located adjacent to, or surrounding, the HA1 amino acid residue at position 162. These portions consisted of the amino acid residues at positions 115–128 (115-QIIPKSSWSDHEAS-128) that is located in the antigenic site A in H3 and at positions 158–170 (158-PTIKRSYNNTNQE-170) that is located in the antigenic site Sa in H1 HA [39] (Fig. 3D). These two domains may affect the binding of the 3B5.1 and 3B5.2 mAbs.
However, because no evidence was found in this study to support 3B5.1 and 3B5.2 mAbs recognizing HA1 amino acids at positions 115–128, the amino acids at positions 158–170, containing the escape-mutant site at position 162, seem to be the epitope recognized by these mAbs. Thus, the HA1 amino acids at positions 158–170 were analyzed to identify the antigenic sites of the 3B5.1 and 3B5.2 mAbs. The residues 158-PTIKRSYNNTNQE-170, present in the HA1 protein of the Ck/Yamaguchi/7/04 strain, were examined to verify whether these residues were conserved among other H5 subtype strains, including the six H5N1 HPAIV strains and seven LPAIV (EA-nonGsGD and Am-nonGsGD clades) strains used in this study (Table 4). The amino acid sequence of the six HPAIV strains was the same, for this region, as the Ck/Yamaguchi/7/04 strain, except at position 162 for clades 2.3.2.1, 2.3.2.1a, and 2.3.2.1c (K162), and at position 163 for clades 2.3.2.1a and 2.3.2.1c (G163). However, the lysine residues at position 162 (K162) did not affect the binding ability of the 3B5.1 and 3B5.2 mAbs; however, the mAbs could not react with mutants R162I or R162G (Fig. 3A).
In addition, five EA-nonGsGD clade strains recognized by the 3B5.1 and 3B5.2 mAbs had the same amino acid sequence (158-PTIKRSYNNTNQE-170) as the Ck/Yamaguchi/7/04 strain, except at position 169 in the Dk/Hong Kong/820/83 strain (Q169K) and at position 170 of the Md/Hokkaido/24/09 strain (E170D) (Table 4). However, these mutated amino acid residues (Q169K and E170D) did not affect the binding ability of 3B5.1 and 3B5.2 mAbs. Unexpectedly, in two Am-nonGsGD strains; the Ck/Ibaraki/1/05 strain which possessed P158K, I160L, S163N, N165T, and Q169V substitutions and the Whis. sw/Shimane/499/83 strain which possessed K161E, S163T, and Q169V substitutions, the HA1 proteins were not recognized by the 3B5.1 and 3B5.2 mAbs.
Conservation of the HA1 epitope (158–170) among divergent H5N1 HPAIVs recognized by the mAbs
The 158-PTIKRSYNNTNQE-170 epitope was aligned and compared to 5,366 strains from clades 0–9 of H5N1 to analyze the conservation of this epitope (Table 5). The result showed that the HA1 amino acid at positions 158, 160, 164, and 166 (P158, I160, Y164, and N166) were highly conserved (>90%) among clades 0–9. R162 and K162 in HA1 were also highly conserved among all clades, except clades 2.2.1.1a (E162), 2.2.2 (I162), 2.2.2.1 (I162), 2.3.1 (I162), 2.3.4.4 (I162), 7.1 (V162) and 7.2 (V162). The amino acid positions 159, 163, 165, and 167–170 (T159, S163, N165, T167, N168, Q169, and E170) were variable in clades 2.3.2.1b (I159 and K169), 7.1 (P159, N163, and T165), 7.2 (P159, N163, T165, and A167), 2.1.3.2a (T163), 2.1.3.2b (T163), 2.3.2.1c (G163), 2.2.1.1 (H165), 2.2.1.1a (H165), 2.1.3 (E168), and 2.3.4.4 (R169) and mutated and moderately conserved (50–90%) in clades 1.1.1 (D168), 2.3.4.1 (S168), 2.3.2.1c (R169) and 7.2 (K170).
Discussion
In this study, three neutralizing anti-H5 mAbs against clades 2.2 (1G5 mAb) and 2.5 (3B5.1 and 3B5.2 mAbs) viruses were successfully established (Fig. 1A). Interestingly, the 3B5.1 and 3B5.2 mAbs reacted to all HPAIV strains from clades 1, 2.2, 2.3.2.1, 2.3.2.1a, 2.3.2.1b, 2.3.2.1c, and 2.3.4 isolated in Japan and Vietnam during 2003–2012, indicating that the mAbs recognized a cross-clade specific epitope; whereas the 1G5 mAb did not react with all the clades examined (Table 2).
Although four amino acid mutations (S56N, R162I, R162G, and A184G, H5 numbering) were found in escape mutants (3B5.1 and 3B5.2 mts) (Fig. 3A), the mutations at positions 56 and 184 did not seem to affect the binding ability of 3B5.1 and 3B5.2 mAbs, since the amino acid at position 56 was buried in the HA1 protein, and clades 2.3.2.1, 2.3.2.1a, and 2.3.2.1c containing E184 (data not shown) were recognized by those mAbs (Table 2). In contrast, the positively charged amino acid residue (R162) seemed to associate highly with the binding ability of 3B5.1 and 3B5.2 mAbs, since the change from a positively charged (R162) to neutral residue (I162 and G162) led to an inhibition in binding (Fig. 3A). However, although H5N1 clade 2.3.2.1, 2.3.2.1a, and 2.3.2.1c strains have a lysine residue at position 162 (K162) and not arginine, they were also recognized by these mAbs. Since the HA amino acid sequences of the clade 2.3.2.1b strains used in this study were not available, we examined the 35 HA sequences of clade 2.3.2.1b strains available in the database. We found that all strains contained the amino acid residue K162, as shown in Table 5. Therefore, the 3B5.1 and 3B5.2 mAbs mainly interact with positively charged amino acid residues at position 162 in the HA1 protein.
The mutation of amino acids at position 162 of the HA1 protein caused a loss in binding of some anti-H5 mouse mAbs [40–42]. The VN04-2 mAb to the A/Vietnam/1203/04 (clade 1) strain reacted to clade 2.1.1 (R162), but not to clade 2.1.3.1, 2.2, and 2.3.4 strains which have the same residue R162 [40]. Four anti-H5 mouse mAbs to Duck/Novosibirsk/56/05 (clade 2.2) (3G9, 5G9, 5F12, and 6E2) seemed to recognize R162, because they failed to react with the escape mutants with the amino acid residue change R162G/K/W; however, these mAbs could not recognize clade 1 (R162), clade 2.3.2.1 (K162) and 2.3.2.1c (K162) strains [41, 42]. On the other hand, our mAbs (3B5.1 and 3B5.2) could recognize clade 1 (R162) as well as clades 2.1.3.1 (R162), 2.2 (R162), 2.3.2.1(K162), 2.3.2.1c (K162), and 2.3.4 (R162). Thus, our mAbs also recognized the H5N1 strains with K162 in the HA1 protein (Table 4), a result that contradicts with previous studies [40–42]. Although the reason for this contradiction is unclear, the antigenic structures of these regions, including the important amino acid residue at position 162 that our mAbs (3B5.1 and 3B5.2) recognize, may be different from those that other mAbs recognize.
Although several, broad, cross-clade reactive anti-H5 HA human and mouse mAbs have been reported [17–20], there is not much information on their reactivity to the newly emerging H5 HPAIVs (clades 2.3.2.1a or 2.3.2.1c). Although a human mAb (FLD194) recognized A/Hubei/01/10 strain (clade 2.3.2.1a; up to date at this time) bearing Q119 within the HA1 protein (H5 numbering), in addition to clade 0, 1, 2.1.3, 2.2, 2.2.1, 2.3.1, 2.3.2, 2.3.4, and 2.5 strains, it could not react to two other strains from the same clade 2.3.2.1a (A/chicken/Bangladesh/11RS1984-33/11 and A/chicken/Bangladesh/14VIR2665-23/14), bearing R119 and K119, respectively [20].
Thus, we may need to examine as many strains as possible from different origins and from within clades to define the broad cross-reactivity of established mAbs. Conversely, our mAbs (3B5.1 and 3B5.2) could react to all six strains of clade 2.3.2.1a strains bearing K119 (Table 2) as well as EA-nonGsGD LPAIV strains bearing R119 (Table 3). Although the clade 2.3.2.1a strains used in the previous study [20] were not available in this study, the two strains with R162 or K162 residues in the HA1 protein seemed to be recognized by the 3B5.1 and 3B5.2 mAbs.
Hu et al. [19] reported that a human mAb (65C6) binds a conformational epitope, comprising amino acid residues at positions 118, 121, 161, 164, and 167 (H5 numbering) in the HA1 protein, that is conserved among clade 0, 1, 3–9, 2.1.3.2, 2.2, 2.2.1, 2.3.2.1, 2.3.4, 2.4, and 2.5 strains, except for clade 7.1. However, amino acid mutations at position 162 did not significantly affect the binding ability of the 65C6 mAb, but did abolish the binding ability of the 3B5.1 and 3B5.2 mAbs used in this study (Fig. 3A). These results suggest that our mAbs recognize different antigenic structures to those recognized by the 65C6 mAb, although their antigenic sites seemed to be structurally similar and proximal.
The cross-clade epitope in the HA1 protein that centers on the R162 residue has been reported by Zuo et al [43]. They suggested that four major vulnerable antigenic sites (VS1–4) exist on the A/Anhui/1/05 HA1 protein based on epitopes recognized by H5-specific human and murine mAbs, through analysis of other reports. One of the mAbs (65C6) recognized a cross-clade conformational epitope in VS1 (115-QIIPKSSWSDH-125 and 158-PTIKRSYNNTNQE-170, H5 numbering) that roughly corresponded to Site A on H3 HA and Sa on H1 HA. However, the 158-PTIKRSYNNTNQE-170 region only includes amino acids at positions 158–163 (H5 numbering) of the Sa antigenic site on H1 HA, but not Site A on H3. In this study, 3B5.1 and 3B5.2 mAbs could not bind to mutant strains with the mutation R162I or R162G, which may suggest that the epitope recognized by these mAbs overlapped this epitope (158-PTIKRSYNNTNQE-170).
Therefore, we tried to analyze whether the 158-PTIKRSYNNTNQE-170 epitope was conserved among the 5,366 sequences of various H5N1 clade HAs currently available in the database. As shown in Table 5, the R162 or K162 residues in the HA1 protein were highly conserved among 36 of the 43 H5N1 clades reported, including clades 2.3.2.1a and 2.3.2.1c,, but not 2.2.1.1a, 2.2.2, 2.2.2.1, 2.3.1, 2.3.4.4, 7.1 and 7.2. Out of the 18 amino acid residues in the epitope, four or five residues (159, 162, 163, 165, and 167) in clades 7.1 and 7.2, showed a very low level of residue conservation when compared with other clades (Table 5). It was expected that our mAbs would fail to react to these strains; thus, we believe that clades of H5N1 HPAIV containing the R162 or K162 residue in the HA1 protein could be recognized by our mAbs (3B5.1 and 3B5.2), leading to the positive detection of clade 2.3.2.1a and 2.3.2.1c viruses currently circulating in Asia, Africa, and Europe [11–13].
As shown in Tables 3 and 4, the 3B5.1 and 3B5.2 mAbs recognized the EA-nonGsGD clade (Eurasian-lineage) H5 LPAIV strains (H5N1, H5N2, and H5N3) with an R162 residue in the HA1 protein. Unexpectedly, these mAbs could not recognize the Am-nonGsGD clade (American-lineage) H5 LPAIV strains (H5N2 and H5N3), even those with the residue R162. In the American-lineage strains, the amino acid residue at position 158 in the HA1 protein of the Ck/Ibaraki/1/05 strain (K158) was positively charged, whereas the residue at position 161 of the Whis.sw./Shimane/499/83 strain (E161) was negatively charged. These positively and negatively charged amino acid residues were different from the Eurasian-lineage strains of H5 HPAIV and LPAIV (P158 and K161) (Table 4). The shift from positive to negative charge has been reported to result in conformational changes on the HA1 surface that affect the binding of anti-H5 mAbs [17]. Herein, the negatively charged amino acid residue (E161) within Whis.sw./Shimane/499/83 may have affected the binding of the 3B5.1 and 3B5.2 mAbs. However, it was unclear whether the positively charged residue (K159) in Ck/Ibaraki/1/05 strain similarly affected the binding of these mAbs. We suspect that the HA1 structure of this Am-nonGsGD clade strain was complex and different from that of the HPAIV and EA-nonGsGD clade strains.
Unfortunately, the 3B5.1 and 3B5.2 mAbs may not recognize clade 2.2.1.1a, 2.2.2, 2.2.2.1, 2.3.1, 2.3.4.4, 7.1, and 7.2 strains, containing the residues: E162, I162, or V162 within the HA1 protein (Table 5). Therefore, in order to detect clades 2.2.1.1a, 2.2.2, 2.2.2.1, 2.3.1, 2.3.4.4, 7.1, and 7.2, additional mAbs should be produced. A combination of our mAbs (3B5.1 and 3B5.2) and the complementary mAbs to clade 2.2.1.1a, 2.2.2, 2.2.2.1, 2.3.1, 2.3.4.4, 7.1, and 7.2 strains could be useful in the development of a diagnostic test with broad cross-clade reactivity for detection of all H5N1 strains. Previously, a mAb-based dot ELISA was developed for the universal detection of H5N1 viruses including clades 0, 1, 2.1, 2.2, 2.3, 4, 7, and 8 by using two complementary anti-H5 mAbs [44].
Outbreaks caused by H5N1 HPAIVs still cause serious problems and have an impact on both animal and public health. Therefore, rapid diagnostic tests are essential for the detection and monitoring of circulating H5N1 viruses and to prevent outbreaks. Our results indicate that the 3B5.1 and 3B5.2 mAbs have potential for this application due to their broad cross-reactivity to H5N1 viruses. These mAbs could be available to develop diagnostic tests for recently circulating H5N1 viruses, especially for clade 2.3.2.1a and 2.3.2.1c strains that have been circulating and causing lethal infections of poultry in Asia, Africa, and Europe [11–13].
References
Chan PK (2002) Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis 34(Suppl 2):S58–S64
Brown IH (2010) Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis 54:187–193
Amendola A, Ranghiero A, Zanetti A, Pariani E (2011) Is avian influenza virus A(H5N1) a real threat to human health? J Prev Med Hyg 52:107–110
OIE and FAO (2015) OFFLU Annual Report 2015. http://www.offlu.net/fileadmin/home/en/publications/pdf/OFFLU_Annual_Report_2015.pdf. Accessed 20 Oct 2016
WHO/OIE/FAO, H5N1 Evolution Working Group (2008) Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 14:e1
WHO/OIE/FAO, H5N1 Evolution Working Group (2014) Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses 8:384–388
WHO/OIE/FAO, H5N1 Evolution Working Group (2012) Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Respir Viruses 6:1–5
Smith GJ, Donis RO, WHO/OIE/FAO, H5 Evolution Working Group (2015) Nomenclature updates resulting from the evolution of avian influenza A (H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir Viruses 9:271–276
WHO (2014) Antigenic and genetic characteristics of zoonotic influenza viruses and candidate vaccine viruses developed for potential use in human vaccines. http://www.who.int/influenza/vaccines/virus/201409_zoonotic_vaccinevirusupdate.pdf. Accessed 20 June 2016
WHO (2015) Antigenic and genetic characteristics of zoonotic influenza viruses and candidate vaccine viruses developed for potential use in human vaccines. http://www.who.int/influenza/vaccines/virus/201502_zoonotic_vaccinevirusupdate.pdf. Accessed 20 June 2016
WHO (2016) Antigenic and genetic characteristics of zoonotic influenza viruses and candidate vaccine viruses developed for potential use in human vaccines. http://www.who.int/influenza/vaccines/virus/201602_zoonotic_vaccinevirusupdate.pdf. Accessed 20 June 2016
Swayne DE (2007) Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis 51(1 Suppl):242–249
Spackman E, Swayne DE, Suarez DL, Senne DA, Pedersen JC, Killian ML, Pasick JK, Handel K, Lee CW, Stallknecht D, Slemons R, Ip HS, Deliberto T (2007) Characterization of low-pathogenicity H5N1 avian influenza viruses from North America. J Virol 81(21):11612–11619
Ohnishi K, Takahashi Y, Kono N, Nakajima N, Mizukoshi F, Misawa S, Yamamoto T, Mitsuki YY, Fu S, Hirayama N, Ohshima M, Ato M, Kageyama T, Odagiri T, Tashiro M, Kobayashi K, Itamura S, Tsunetsugu-Yokota Y (2012) Newly established monoclonal antibodies for immunological detection of H5N1 influenza virus. Jpn J Infect Dis 65:19–27
Du A, Daidoji T, Koma T, Ibrahim MS, Nakamura S, de Silva UC, Ueda M, Yang CS, Yasunaga T, Ikuta K, Nakaya T (2009) Detection of circulating Asian H5N1 viruses by a newly established monoclonal antibody. Biochem Biophys Res Commun 378:197–202
Wada A, Sakoda Y, Oyamada T, Kida H (2011) Development of a highly sensitive immunochromatographic detection kit for H5 influenza virus hemagglutinin using silver amplification. J Virol Methods 178:82–86
Kobayashi-Ishihara M, Takahashi H, Ohnishi K, Nishimura K, Terahara K, Ato M, Itamura S, Kageyama T, Tsunetsugu-Yokota Y (2014) Broad cross-reactive epitopes of the H5N1 influenza virus identified by murine antibodies against the A/Vietnam/1194/2004 hemagglutinin. PLoS One 9:e99201
Wu R, Li X, Leung HC, Cao Z, Qiu Z, Zhou Y, Zheng BJ, He Y (2014) A novel neutralizing antibody against diverse clades of H5N1 influenza virus and its mutants capable of airborne transmission. Antiviral Res 106:13–23
Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, Calder L, Gamblin SJ, Zhang L, Deubel V, Zhou B, Skehel JJ, Zhou P (2012) A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol 86:2978–2989
Xiong X, Corti D, Liu J, Pinna D, Foglierini M, Calder LJ, Martin SR, Lin YP, Walker PA, Collins PJ, Monne I, Suguitan AL Jr, Santos C, Temperton NJ, Subbarao K, Lanzavecchia A, Gamblin SJ, Skehel JJ (2015) Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci U S A 112:9430–9435
Imai K, Ogawa H, Bui VN, Inoue H, Fukuda J, Ohba M, Yamamoto Y, Nakamura K (2012) Inactivation of high and low pathogenic avian influenza virus H5 subtypes by copper ions incorporated in zeolite-textile materials. Antiviral Res 93:225–233
Trinh DQ, Ogawa H, Bui VN, Baatartsogt T, Kizito MK, Yamaguchi S, Imai K (2015) Characterization of mAbs to chicken anemia virus and epitope mapping on its viral protein, VP1. J Gen Virol 96:1086–1097
Harlow E, Lane D (1988) Antibodies A Laboratory Manual. Cold Spring Harbor Laboratory, New York
Nishikawa K, Isomura S, Suzuki S, Watanabe E, Hamaguchi M, Yoshida T, Nagai Y (1983) Monoclonal antibodies to the HN glycoprotein of Newcastle disease virus. Biological characterization and use for strain comparisons. Virology 130:318–330
WHO (2005) WHO Manual on Animal Influenza Diagnosis and Surveillance http://apps.who.int/iris/bitstream/10665/68026/1/WHO_CDS_CSR_NCS_2002.5.pdf. Accessed 20 June 2016
Behrens B, Kärber G (1934) Wie sind Reihenversuche für biologishe Auswertungen am zweckmäßigsten anzuordnen [in German]? Naunyn Schmiedebergs. Arch Exp Pathol Pharmakol 18:379–388
Masuda T, Nagai M, Yamasato H, Tsuchiaka S, Okazaki S, Katayama Y, Oba M, Nishiura N, Sassa Y, Omatsu T, Furuya T, Koyama S, Shirai J, Taniguchi K, Fujii Y, Todaka R, Katayama K, Mizutani T (2014) Identification of novel bovine group A rotavirus G15P[14] strain from epizootic diarrhea of adult cows by de novo sequencing using a next-generation sequencer. Vet Microbiol 171:66–73
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols 10:845–858
DeLano WL, Bromberg S (2004) PyMOL User’s Guide. http://pymol.sourceforge.net/newman/userman.pdf. Accessed 20 June 2016
Mase M, Eto M, Tanimura N, Imai K, Tsukamoto K, Horimoto T, Kawaoka Y, Yamaguchi S (2005) Isolation of a genotypically unique H5N1 influenza virus from duck meat imported into Japan from China. Virology 339:101–109
Suzuki K, Okada H, Itoh T, Tada T, Tsukamoto K (2010) Phenotypes influencing the transmissibility of highly pathogenic avian influenza viruses in chickens. J Gen Virol 91:2302–2306
Bui VN, Ogawa H, Ngo LH, Baatartsogt T, Abao LN, Tamaki S, Saito K, Watanabe Y, Runstadler J, Imai K (2013) H5N1 highly pathogenic avian influenza virus isolated from conjunctiva of a whooper swan with neurological signs. Arch Virol 158:451–455
Okamatsu M, Tanaka T, Yamamoto N, Sakoda Y, Sasaki T, Tsuda Y, Isoda N, Kokumai N, Takada A, Umemura T, Kida H (2010) Antigenic, genetic, and pathogenic characterization of H5N1 highly pathogenic avian influenza viruses isolated from dead whooper swans (Cygnus cygnus) found in northern Japan in 2008. Virus Genes 41:351–357
Yamamoto N, Sakoda Y, Motoshima M, Yoshino F, Soda K, Okamatsu M, Kida H (2011) Characterization of a non-pathogenic H5N1 influenza virus isolated from a migratory duck flying from Siberia in Hokkaido, Japan, in October 2009. Virol J 8:65
Okamatsu M, Saito T, Yamamoto Y, Mase M, Tsuduku S, Nakamura K, Tsukamoto K, Yamaguchi S (2007) Low pathogenicity H5N2 avian influenza outbreak in Japan during the 2005-2006. Vet Microbiol 124:35–46
Eto M, Mase M (2003) Isolation of the newcastle disease virus and the H9N2 influenza A virus from chicken imported from China. J Japan Vet Med Assoc 56:333–339
Rudneva IA, Kushch AA, Masalova OV, Timofeeva TA, Klimova RR, Shilov AA, Ignatieva AV, Krylov PS, Kaverin NV (2010) Antigenic epitopes in the hemagglutinin of Qinghai-type influenza H5N1 virus. Viral Immunol 23:181–187
Shore DA, Yang H, Balish AL, Shepard SS, Carney PJ, Chang JC, Davis CT, Donis RO, Villanueva JM, Klimov AI, Stevens J (2013) Structural and antigenic variation among diverse clade 2 H5N1 viruses. PLoS One 8:e75209
Yang H, Carney PJ, Mishin VP, Guo Z, Chang JC, Wentworth DE, Gubareva LV, Stevens J (2016) Molecular characterizations of surface proteins hemagglutinin and neuraminidase from recent H5Nx avian influenza viruses. J Virol 90:5770–5784
Kaverin NV, Rudneva IA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, Krylov PS, Webster RG (2007) Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J Virol 81:12911–12917
Masalova OV, Klimova RR, Chichev EV, Fediakina IT, Loginova SY, Borisevich SV, Bondarev VP, Deryabin PG, Lvov DK, Kushch AA (2011) Development of monoclonal antibodies to highly pathogenic avian influenza H5N1 virus and their application to diagnostics, prophylaxis, and therapy. Acta Virol 55:3–14
Rudneva IA, Kushch AA, Masalova OV, Timofeeva TA, Klimova RR, Shilov AA, Ignatieva AV, Krylov PS, Kaverin NV (2010) Antigenic epitopes in the hemagglutinin of Qinghai-type influenza H5N1 virus. Viral Immunol 23:181–187
Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, Shi X, Liu X, Fan S, Ren H, Hu H, Sun L, Zhou B, Liang M, Zhou P, Wang X, Zhang L (2015) Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun 6:8855
He F, Soejoedono RD, Murtini S, Goutama M, Kwang J (2010) Complementary monoclonal antibody-based dot ELISA for universal detection of H5 avian influenza virus. BMC Microbiol 10:330
Acknowledgements
This work was partially supported by JSPS KAKENHI Grant Number 15H05260, and a Grant-in-Aid for the Bilateral Joint Projects of the JSPS. We would like to thank the staff of National Institute of Veterinary Research in Vietnam to conduct the HI tests and Ms. Sachiko Matsuda in our laboratory for her excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gronsang, D., Bui, A.N., Trinh, D.Q. et al. Characterization of cross-clade monoclonal antibodies against H5N1 highly pathogenic avian influenza virus and their application to the antigenic analysis of diverse H5 subtype viruses. Arch Virol 162, 2257–2269 (2017). https://doi.org/10.1007/s00705-017-3350-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3350-0