Abstract
Endemic genera on oceanic islands often evolved striking morphological and ecological differences among species, with weak postzygotic reproductive isolation. Human activities can lead to increased connectivity and can thereby promote secondary contact and hybridization between previously isolated species. We studied this phenomenon in three species of the genus Pericallis (Asteraceae) on Tenerife, Canary Islands. A total of 53 populations was sampled, including morphologically uniform as well as mixed populations containing morphologically diverse individuals. All plants were analyzed both genetically using AFLP markers and morphometrically. As expected, morphological analysis clearly separated the plants from the uniform populations in three clusters, with distinguishing characters corresponding to those used in species identification. The three species were also grouped into distinct genetic clusters in a structure analysis, although no private alleles were observed. Adding the mixed population data to the analyses provided evidence for extensive hybridization among species, predominantly between P. cruenta and P. echinata, and between P. cruenta and P. tussilaginis, and morphological and genetic signals were congruent. The mixed populations were geographically located in between the uniform populations and were found in road verges significantly more often than the uniform populations. The observed distribution strongly suggests that hybridization between originally isolated species is recent, promoted by secondary contact due to human disturbance and the construction of roads.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Humans have a strong effect on evolution through their influence on the landscape and the distribution patterns of species. The consequences of human actions on landscape connectivity are twofold. On the one hand, natural habitats are increasingly being fragmented, which negatively affects the population viability of many species (e.g., Young and Clarke 2000; Oostermeijer et al. 2003). On the other hand, natural barriers that originally played an important role in speciation processes are being broken down by the creation of infrastructure (roads, waterways), so that previously isolated sister species can hybridize after establishing secondary contact (Rhymer and Simberloff 1996; Francisco-Ortega et al. 2000). There are several examples of such anthropogenic hybridization. In Australia, disturbed habitats such as road verges bring together previously isolated Banksia (Proteaceae) species and disrupt phenological barriers, promoting hybridization (Lamont et al. 2003). On Tenerife, Argyranthemum frutescens and A. coronopifolium (Asteraceae) form hybrids along roadsides and following the creation of tunnels connecting previously isolated regions (Brochmann 1984). Extensive systems of water courses linking previously separated headwaters have lead to bidirectional introgression in galaxiid fishes in New Zealand (Esa et al. 2000). Man-made landscapes can also provide suitable novel habitats for the establishment of newly formed hybrids. In Switzerland, Cardamine insueta (Brassicaceae), originating from hybridization between C. rivularis and C. amara, especially invaded disturbed hay meadows (Urbanska et al. 1997). In Germany, hybridization among species of Rorippa (Brassicaceae) occurs predominantly in man-made habitats such as drainage ditches (Bleeker and Hurka 2001). In addition to directly disrupting natural barriers, humans also increasingly transport species across them (e.g., Hodkinson and Thompson 1997; von der Lippe and Kowarik 2007; Wichmann et al. 2009), resulting in problems of invasiveness that frequently involve hybridization between native and non-native (or genetically modified crop) species (Schierenbeck and Ellstrand 2009).
Oceanic islands are well known for harboring large numbers of, often closely related, endemic species that are the result of adaptive radiation (Schluter 2000). Due to the specific geographical histories of the oceanic archipelagos, island endemics are often highly specialized and have extremely narrow geographic distributions (Baldwin et al. 1998; Francisco-Ortega et al. 2000; Emerson 2002). Unfortunately, a high proportion of these species is threatened or has already gone extinct because of anthropogenic activities. Introduction of non-native species, habitat destruction through deforestation and development of human infrastructure are all well known to have a major impact on native island biodiversity (Whittaker and Fernández-Palacios 2007). Because of their often recent evolutionary origin, island endemics tend to hybridize easily despite morphological and ecological differences (Baldwin et al. 1998; Francisco-Ortega et al. 2000; Fjellheim et al. 2009). Postzygotic barriers are typically weak (Carlquist and Cole 1974; Levin et al. 1996), and hybrids are often fertile so that introgression can readily occur. The construction of roads can have a particularly strong influence since they can connect previously isolated populations and the road verges can provide a habitat for the newly formed hybrids (Levin et al. 1996). The combination of the large impact of human activities on oceanic islands and the weak reproductive barriers between species makes island endemics especially vulnerable to human-induced hybridization.
A genus that is potentially threatened by human-induced hybridization is Pericallis D. Don in Sweet (Asteracae, Senecioninae), whose distribution is limited to the Macaronesian archipelago. On the Canary island of Tenerife, three herbaceous species occur that originally exhibited geographically or ecologically isolated distributions: P. echinata (L. fil.) B. Nord., P. cruenta (L’Her.) Bolle and P. tussilaginis (L’Her.) D. Don (Hohenester and Welss 1993; Schönfelder and Schönfelder 1997; Bramwell and Bramwell 2001). At present, however, many Pericallis populations contain morphological intermediates among these three species. Swenson and Manns (2003) describe morphological intermediates between P. cruenta and P. echinata, although there was no genetic verification of their hybrid status since they did not include these morphological intermediates in their phylogenetic study of the genus. Furthermore, it remains unknown how the human activities on Tenerife have contributed to the hybridization between these Pericallis species.
In this study, we investigate morphological and genetic differentiation among populations of P. echinata, P. cruenta and P. tussilaginis across their main distribution on Tenerife. Using this information, we assess where and to what extent hybridization among these species has occurred. Specifically, we ask:
-
1.
To what extent are the three species morphologically and genetically distinct?
-
2.
Are plants that are morphologically intermediate or exhibit mixed characteristics indeed hybrids and therefore also genetically intermediate?
-
3.
Can we use genetics to detect the geographic regions in which the three species originally evolved?
-
4.
Are the hybrids geographically positioned in between the distribution areas of their parental species, and do they indeed occur more frequently in road verges?
Materials and methods
Study species
The genus Pericallis comprises 14 species (Nordenstam 1978) and has been estimated to be 12–14 million years old (Panero et al. 1999; Swenson and Manns 2003). Here, we study three Pericallis species occurring on the island of Tenerife, P. echinata, P. cruenta and P. tussilaginis. P. cruenta is also found on La Gomera, Gran Canaria and El Hierro, while P. tussilaginis is found on Gran Canaria. P. echinata is endemic to Tenerife (Gobierno de Canarias 2011). Based on ITS sequences, it was shown that P. echinata and P. cruenta belong to a well-supported subgroup consisting only of herbaceous species. P. tussilaginis belongs to a sister group that also contains some woody species (Panero et al. 1999; Swenson and Manns 2003). The species descriptions in recent floras (Hohenester and Welss 1993; Schönfelder and Schönfelder 1997; Bramwell and Bramwell 2001) are mainly based on the system of Schultz (1845–1846). This comprises differences in flower disc, leaf and stem color, the presence of spines on the involucral bracts and several leaf characteristics (Table 1).
On Tenerife, the three species have limited, but partly overlapping distributions. Both P. echinata and P. tussilaginis are described as typical for the (former) laurel forest and its margins, although they extend upward into the pine forest belt and downward into the lowland succulent shrub zone (Table 1). Pericallis cruenta is characteristic for pine forests (Hohenester and Welss 1993; Schönfelder and Schönfelder 1997; Panero et al. 1999; Bramwell and Bramwell 2001). In addition to their occurrence in natural areas, all three species are frequently found in road verges and the margins of small arable fields (Rivas-Martínez et al. 1993). Natural population sizes may range from a few up to several hundreds of individuals, while linear populations in road verges may contain thousands of plants. All species are diploid and have a relatively high chromosome number (2n = 60, Suda et al. 2003, 2005). Pollination experiments in our greenhouse have showed that all three species are predominantly outcrossing (R. Bregman and J.C.M. den Nijs, unpublished data). The flowering time of the three study species is partly overlapping (Table 1).
Sampling
From the island of Tenerife, we sampled a total of 53 Pericallis populations. Of these, 51 were sampled along a belt transect covering the north side of the island from east to west, and from low to high elevations. In addition, two isolated populations of putative P. echinata were sampled in the SW region around Roque del Conde. Together, the sampling sites covered most of the known range of the three species. Sampling sites covered various vegetation zones, the succulent scrub zone (0–700 m), the evergreen forest zone (400–1,500 m) and the lower pine forest zone (1,200–2,000 m; Bramwell and Bramwell 2001) and completely natural as well as human-disturbed habitats. The choice of sampling locations was partly guided by distributional information obtained from herbarium specimens (Universidad de la Laguna, herbarium TFC). The herbarium specimens were also used to familiarize ourselves with the morphology of the three species and to aid in the selection of diagnostic morphological characters. The exact locations of the sampling sites and whether or not they were located in a road verge are listed in Table S1 (online supplementary material).
In each population we first surveyed the morphological range that was present. We then collected five individuals that represented the morphological variation of the population to maximize variability in our sample. Plants were identified in the field using the species’ descriptions and keys of the Floras of Tenerife (Table 1). On the basis of these identifications, a population was labeled as “uniform” when all plants belonged to a single taxon or “mixed” when not all plants could be unambiguously assigned to one of the three species. A sample of young leaf material was stored in a small zip-loc bag with silica gel for later DNA extraction and AFLP analysis. In total, we sampled 265 individuals from 53 populations. The series of populations comprised 11 uniform populations of P. tussilaginis, 11 of P. cruenta and 15 of P. echinata. Sixteen “mixed” populations contained one or more individuals with character states that were intermediate between two or three of the study species.
Morphological analysis
We used a set of 17 morphological variables, including 8 floral and 9 vegetative characters (Table 2; Fig. 1). The number of flower heads was counted from above the last node with an attached leaf of >1 cm. The color of the ray flowers was determined using the color book by Kornerup and Wanscher (1967). Numerical flower traits (diameter of the flower disc, length of the ray flowers, width of the ray flowers and number of spines per involucral bract) were based on the average of measurements on five flower heads randomly selected from the inflorescence. Leaf characters were measured on the fifth leaf, counting from the tip of the inflorescence. Stem characteristics were measured at the position where this fifth leaf was attached. Three primary measurements were transformed to ratios (see legend Fig. 1) to obtain shape estimates: heart shape of the leaf blade, relative auricle size and auricle shape (Fig. 1). A stereomicroscope was used for counting the number of spines per involucral bract and the number of hairs per unit surface on the upper side of the leaf. In order to normalize the data, some of the morphological variables were log-transformed.
DNA isolation
Total plant DNA was extracted from approximately 15 mg dried leaf material using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1987). DNA samples that resulted in poor preamplification DNA were isolated a second time using an Invitek Invisorb Spin Plant Mini Kit. DNA concentrations were measured on a NanoDrop ND-1000 spectrophotometer and standardized in TE0.1 20 ng/µl.
AFLP analysis
We used AFLP analysis (Vos et al. 1995) for genotyping all individuals. Though AFLPs are dominant anonymous markers and therefore have some drawbacks, they have been successfully used to detect hybridization and admixture (Blair and Hufbauer 2010; Mráz et al. 2011). AFLPs are a good choice for taxa like Pericallis for which little prior genomic information is available (Bonin et al. 2007). The AFLPs were performed using the Invitrogen AFLP Analysis System I and Invitrogen AFLP Starter Primer Kit. However, we slightly changed the protocol provided by the company as follows. Ligation took place at room temperature for 1 h. Afterwards the samples were placed overnight in a water bath gradually decreasing from room temperature to 4 °C. PCR reactions were carried out in a PTC 200 thermocycler (MJ Research). Preselective amplification consisted of 24 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s, followed by 5 min at 72 °C and 5 min at 4 °C. The PCR product was diluted 50 times in TE0.1 buffer. The preamplification product was loaded onto an agarose gel to assess its quality. The PCR for selective amplification contained 1 × PCR buffer [HT Biotechnology: 100 mM Tris-HCl, pH 9.0, 15 mM MgCl2, 500 mM KCl, 1 % Triton X-100, 0.1 % (w/v) stabiliser], 0.1 mM of each dNTP, 0.15 µM of each primer and 0.04 units/µl Taq DNA polymerase (HT Biotechnology) in 10 µl reaction volume. The PCR of the selective amplification was hot-started at 94 °C for 1 min, followed by 12 cycles starting with 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 60 s and stepping down the annealing temperature by 0.7 °C every cycle, and another 24 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, again finishing with 5 min at 72 °C and 5 min at 4 °C.
From 18 tested primer combinations we selected two pairs (MSE-CAT/Eco-AGC and MSE-CAT/Eco-ATC) that gave sufficient variation between the samples and a suitable number of well-separated fragments. The number of markers was 185 in MSE-CAT/Eco-AGC and 179 in MSE-CAT/Eco-ATC. We selected 35 markers of MSE-CAT/Eco-AGC and 18 of MSE-CAT/Eco-ATC that were polymorphic and could be readily identified on all gels. We separated the selective amplification products on a Gene ReadIR 4200 Analyzer (Li-Cor, Inc.).
We minimized genotyping errors by adhering to the recommendations given by Bonin et al. (2004). For instance, the AFLP fragments were visually scored as present (1) or absent (0) independently by two persons. Furthermore, low quality DNA samples and weakly amplified samples were discarded; the number of successfully genotyped individuals per population is listed in Table S1 (online supplementary material). The AFLP matrix has been published in the Demiurge database (http://www.demiurge-project.org/matrix_digests/30).
Statistical analysis
Morphological variation was analyzed by means of principal component analysis (PCA) based on a correlation matrix, using SPSS 11.0 for Windows. To test the robustness of the PCA to the inclusion of ordinal variables (see Table 2), we performed the PCA both with and without those variables included. Both analyses showed the same general pattern, so we only present the analysis with both ordinal and continuous characters included. We performed a principal coordinate analysis (PCoA) on the AFLP data using the program PAST (Hammer et al. 2001), based on the Gower similarity index (Gower 1966). Genetic differentiation among the uniform populations was calculated for all three species separately and among species in a hierarchical analysis with the populations clustered within species. We calculated both F ST and F’ ST , which is a standardized analogue of F ST that is independent of the within-population diversity (Meirmans and Hedrick 2011). The values of the F-statistics were calculated using the program Genodive (Meirmans and Van Tienderen 2004), and the significance was tested using 999 permutations.
To analyze the kinship of the three species and to quantify the ancestry of the putative hybrids we ran the admixture model of the program structure 2.2 (Pritchard et al. 2000; Falush et al., 2003), using 53 polymorphic markers following the recommendations for dominant markers from Falush et al. (2007). Although structure is best known for its ability to discern the number of population clusters (but see Kalinowski 2011), it can also be used as a powerful tool to detect admixture when the number of species is known in advance (Vaha and Primmer 2006; Thompson et al. 2010), as is the case here. Since three species have been described we started by running the analysis with K = 3 using the AFLP data of the uniform populations only to see if the morphologically defined species were also genetically separated. We set the iteration and start parameters to their default values (with a burnin of 100,000 steps followed by a Monte Carlo Markov chain of 100,000 steps), and used the admixture model without providing any a priori information on the origin of the samples. Since the taxonomic status of the two isolated Roque del Conde populations from the Southwest was uncertain, we excluded these from the initial structure analysis. We tested whether these populations were identified as a distinct cluster by setting K = 4, again without providing a priori information on population or species. Finally, to identify genetically intermediate individuals in mixed populations we again employed the admixture model with K = 4, but now providing the program a priori with information on the three clusters identified in the first step, i.e., the three uniform populations (using the Popinfo setting). No information was given to the program on the identity of plants collected in mixed populations or the Roque del Conde individuals following the approach to identify hybrids in a simulation study of Vaha and Primmer (2006). The results from structure were verified by clustering the data using spatial Bayesian clustering implemented in Tess (François et al. 2006) and the AMOVA based clustering implemented in Genodive (Meirmans and Van Tienderen 2004), which identified the same population clusters (results not shown).
The clustering of individuals based on the AFLP data using structure was compared to the results of a clustering analysis of the morphological data using the fuzzy c-means algorithm (Dunn 1973). Fuzzy c-means is a clustering technique in which every individual can belong to one or more clusters. The results are comparable to the results of a structure analysis, but where structure uses an explicit genetic model, the fuzzy c-means algorithm uses a model-free least-squares approach that makes it suitable for multiple types of data. For fuzzy c-means clustering of the morphological data set, the individuals were clustered into three groups. For this analysis we first removed the individuals from the Roque del Conde populations that in the structure analysis also formed a separate group. We used the characters from Table 2 with the exception of the shape of the spines that was the only non-ordinal categorical trait. The clustering was performed in the statistical framework R (R Development core team, 2010), using the function “cmeans” from the “e1071” package (Dimitriadou et al. 2010). Pearson correlations (one-sided) were calculated among genetic and morphological assignments for each of the three species.
All sampling sites were classified into those that were located in the verge of a paved road and those that were not. Two by two contingency tables were created to compare the frequencies of road verge locations among the mixed and the uniform populations. Significance of the contingency tables was tested using a Fisher exact test, with a sequential Bonferroni correction for multiple testing.
Results
Morphology: principal component analysis
We found three clearly separated groups for P. echinata, P. cruenta and P. tussilaginis. In contrast, the individuals from “mixed” populations form a nearly continuous pattern of variation within and between the three clusters (Fig. 2a). The first principal component separated P. tussilaginis and P. echinata from P. cruenta. Characters contributing to this axis were the color of the underside of the leaves and the flower discs, the hairiness of the stem and leaves, the number of flower heads and the diameter of the flower disc (Table 2). The second axis separated P. echinata from P. cruenta and P. tussilaginis, on the basis of the number of spines on the involucral bracts, the shape of the spines and the relative auricle size (Table 2). Subsequent axes provided no useful information regarding the differences among the species (data not shown). The Roque del Conde individuals were placed within the P. echinata cluster and between the P. echinata and the P. tussilaginis clusters (Fig. 2a).
a Principal component analysis based on 17 morphological characters from Pericallis echinata, P. cruenta and P. tussilaginis. b Principal coordinate analysis based on 53 AFLP markers. Each dot represents an individual; the color labels represent the identification of the population types given at the time of sampling
AFLP data: principal coordinate analysis
None of the individual AFLP markers was diagnostic for a single species, and 38 of the markers (72 %) were polymorphic in all three study species. Five markers (9 %) were never found in P. cruenta, and one marker (2 %) was never found in P. echinata. Furthermore, nine markers (17 %) were always present in P. cruenta.
The PCoA based on 53 AFLP markers (Fig. 2b) is largely similar to the morphological analysis (Fig. 2a). Three groups can clearly be identified for the uniform populations, showing separation of the unambiguously identified individuals of P. echinata, P. cruenta and P. tussilaginis. However, compared to the PCA, the variation within P. echinata in the PCoA is considerably larger.
Inference of population structure
The analysis of the uniform populations showed indeed that the morphological species were also genetically well separated (Fig. 3a), with only a few exceptions for which the genetic assignment differed from the identification based on morphological measurements. The first cluster predominantly contained plants from the uniform P. echinata populations, the second consisted of all P. cruenta populations and the third of all P. tussilaginis populations. The clusters representing P. cruenta and P. tussilaginis were more homogeneous than that of P. echinata, which showed more variation among plants in their assignment to one of the three clusters. The separation of the three species was confirmed using F-statistics; in a hierarchical analysis where populations were grouped within species, the among-species differentiation was strong with a F CT value of 0.182 (P = 0.001, 999 permutations). However, as shown in Table 3, there was also significant differentiation within species for P. cruenta (F ST = 0.204, P = 0.001) and P. echinata (F ST = 0.073, P = 0.001), but not for P. tussilaginis (F ST = 0.034, P = 0.062).
Bayesian assignment of individuals to three (a) and four (b and c) clusters by structure. Each bar represents one individual plant. Individuals are ordered by sampling site, using the morphologically inferred taxonomic status of the populations: (1) uniform populations of Pericallis echinata, (2) uniform P. cruenta, (3) uniform P. tussilaginis, (4) Roque del Conde plants and (5) mixed populations with Pericallis echinata, P. cruenta and tussilaginis characters. For plots a and b, no a priori information was used on the identity of the species. For plot c the identities of cluster 1–3 were given using popinfo option. The colors represent structure’s assignment of individual plants to the different clusters. The dark blue cluster assignment is associated with identification as P. echinata, green with P. cruenta and light blue with P. tussilaginis (all plots), yellow to the Roque del Conde individuals (plot b and c)
To investigate the status of the Roque del Conde populations, the second analysis was performed with these putative P. echinata populations added to the data set, setting the number of clusters to K = 4 (Fig. 3b). The genetic constitution of the Roque del Conde populations appeared to be sufficiently unique to lead to a separate cluster in the analysis, although there is some overlap with the other sites of P. echinata.
The final structure analysis (Fig. 3c) concentrated on the identification of the genetic makeup of plants in mixed populations. By using a priori information on the identity of the three different species in the uniform populations (using the popinfo option in structure) the position of individuals of the mixed populations could be assessed (cf. Vaha and Primmer 2006). The Roque del Conde individuals again surfaced as a separate cluster in this analysis. Within the mixed populations we found admixed as well as genetically pure individuals. The admixed individuals were genetic mixtures of the parental species P. echinata/cruenta and P. cruenta/tussilaginis. A few individuals were complex hybrids containing genetic material from all three parental species. Thus, mixed populations can contain both plants from the different parental species as well as intermediate, putative hybrid genotypes (Fig. 3c).
The morphological fuzzy c-means clustering analysis assigns each sampled individual a probability of belonging to each cluster (representing a species). The scores were highly significantly correlated with those of the structure analysis for all three species (Fig. 4a–c). When the analysis was performed on data of the suspected hybrids only (df = 66), the correlation coefficients were lower, but still highly significant (R² = 0.40, P < 0.001, R² = 0.57, P < 0.001 and R² = 0.40, P < 0.001 for P. echinata, P. cruenta and P. tussilaginis, respectively). This analysis demonstrates that the hybrid indices based on morphological and genetic data are mostly congruent. Despite this congruence, there were some individuals, especially in P. echinata, that were indicated as admixed by structure, but were recorded as morphologically pure. However, when the population assignment was used as priors in the Bayesian analysis, those individuals were also genetically pure (Fig. 3c).
Relationship between the morphological and genetic cluster assignment. The analysis, tested with Pearson’s correlation coefficient (df = 192), shows that plants from uniform populations cluster in two corners: the upper right corner of each graph contains individuals with high genetic and morphological assignment scores for a P. echinata, b P. cruenta and c P. tussilaginis; the lower left corner of each diagram comprises the individuals with low genetic and morphological assignment scores, i.e., from uniform populations of the other species. Individuals scoring intermediately for the genetic as well as morphological clustering analyses mostly come from ‘mixed’ populations (open symbols). All correlations remained significant (P < 0.001) when only the individuals from mixed populations were considered (df = 66)
Location and disturbance of mixed populations
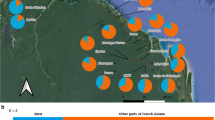
Placement of the results of the structure analysis on the geographical map of Tenerife visualizes the core distribution areas of the three study species and the hybrid zones (Fig. 5). The P. echinata core area is located in the Teno mountains in the NW region of Tenerife, between 50 and approximately 900 m elevation. The core area of P. cruenta is located in the forest zones between approximately 700 and 1,500 m in the N central region. The P. tussilaginis core area is located in the Anaga mountain range in the NE of the island, between approximately 200 and 800 m. In between these core distribution areas, we mostly find mixed populations, generally between 500 and 1,000 m above sea level. Some populations of P. tussilaginis and P. echinata occur beyond the species’ core distribution areas (e.g., populations 16 and 20, and 34 and 38, respectively).
Geographical map of Tenerife showing the approximate sampling locations and the population mean of the Bayesian assignment scores assuming the existence of four clusters, calculated with structure on the basis of AFLP data (see Fig. 3c). The arrows indicate the position of the core distribution areas of Pericallis echinata, P. cruenta and P. tussilaginis according to Bramwell and Bramwell (2001). Grey circles outline the Teno, Anaga and Roque del Conde mountain ranges. Sample numbers above or below each bar correspond with the numbers in Table S1 (online supplementary material)
The mixed populations were more often found in road verges than the uniform populations (Fisher's exact test P = 0.0021). Of the mixed populations 88 % (14 out of 16) were located in road verges, whereas of the uniform populations on average only 40 % were found in road verges (14 out of 35). We also compared the frequency of road verge populations of the mixed and uniform populations separately for the three species. There was a significant difference between the 16 mixed populations and the P. echinata populations (excluding the 2 Roque del Conde populations; 5 out of 13; 38 %; P = 0.016) and also between the mixed and P. cruenta populations (2 out of 11; 18 %; P < 0.001). However, there was no significant difference between the mixed populations and the P. tussilaginis populations (7 out of 11; 64 %; P = 0.19).
Discussion
Hybridization in Pericallis
The clearly separated morphological clusters we found for P. echinata, P. tussilaginis and P. cruenta were in accordance with the results from the genetic analyses; the PCoA, structure analysis and F-statistics all show a high degree of differentiation among the three species. Individuals originating from mixed populations contained pure as well as morphologically and genetically intermediate plants, and were accordingly distributed within and between the clusters of the putative parental species. The presence of morphological and genetic intermediates between P. echinata and P. cruenta and between P. cruenta and P. tussilaginis supports the hypothesis that there is ongoing hybridization between these species. This has been confirmed by experimental crosses between the species where they were fully interfertile (R. Bregman and J. C. M. den Nijs, unpublished data). The admixture analysis using structure showed that the mixed populations comprised individuals with varying degrees of admixture between parent species. These populations contained not only putative F1 individuals that showed approximately 50 % admixture between two species, but also backcrossed individuals and even some individuals that showed admixture between all three parental species. Such complex patterns of hybridization are usually interpreted as evidence that hybridization has been going on for several generations (Lepais et al. 2009; Thompson et al. 2010).
In our morphometric analysis, the Roque del Conde individuals previously ascribed to P. echinata were clearly different from the plants from the P. echinata core area. Morphologically, these individuals were intermediate between the P. echinata and P. tussilaginis clusters. For example, these plants had the spines on the involucral bracts typical for P. echinata, but also leaf auricles similar to the ones of P. tussilaginis. The genetic support for this is mixed, and we can only speculate about the evolutionary origin of these populations. In the genotypic assignment by structure with four clusters they were not admixed, but rather formed their own distinct cluster. This suggests that they may represent a distinct taxon, possibly with an ancestral status since the Roque del Conde region belongs to the geologically oldest part of Tenerife (discussed below). On the other hand, when structure was run with three clusters, the populations were admixed with a 53 % contribution of P. echinata and a 29 % contribution of P. tussilaginis (results not shown). This suggests that they may represent an isolated lineage derived from P. echinata, possibly introgressed with P. tussilaginis, that has experienced genetic drift in the absence of (further) gene flow. Therefore, the exact taxonomic status of the Roque del Conde populations remains to be settled.
Hybridization and anthropogenic disturbance
Our analyses revealed that mixed populations were more often found in road verges than uniform populations. Roads are known to promote hybridization by both serving as corridors for seed and pollen dispersal and by providing open habitat for the newly formed hybrids to colonize (Anderson 1948; Levin et al. 1996). On the Canary Islands, the infrastructure has radically improved since the 1950s (Pott et al. 2003). Currently, the Canary Islands have the highest road density of all the European oceanic islands (6 km km−2), which strongly affects the remaining laurel and pine forest ecosystems (Delgado et al. 2007). However, the relationship between the mixed populations and the road verges was not absolute; some mixed populations were found outside road verges in other types of disturbed habitat such as arable fields. Conversely, uniform populations, especially for P. tussilaginis, were also found in road verges.
The pattern that emerges is that within the three core areas almost all populations are pure, even the ones located in road verges. In between those core areas, mixed populations are more frequent and are mostly located in road verges. This pattern strongly suggests that the construction of the roads gave rise to a secondary contact between the three core areas, resulting in a hybrid swarm of the three species. We hypothesize that P. cruenta used to be geographically and ecologically separated from P. echinata and P. tussilaginis by its occurrence in the pine forest zone at higher elevations. The population structure of P. cruenta was also clearly different from that of the other two species indicated by a much higher value of F ST . Still, the value of 0.204 that was found for P. cruenta was lower than the average value of 0.281 that was found over multiple species of the Canarian flora (Francisco-Ortega et al. 2000; but see Caujapé-Castells 2010). This may be explained by the fact that wind dispersal of seeds is more restricted in the forest habitat of P. cruenta than in the more open habitats of P. echinata and P. tussilaginis. Road verges through forests and deforested areas open up the landscape and provide suitable habitat for P. echinata, allowing it to extend its range upwards and overlap with that of P. cruenta. The human-disturbed areas are colonized by P. cruenta × P. echinata hybrids from Teno to N central Tenerife and P. cruenta × P. tussilaginis hybrids from N central Tenerife to Anaga. On Tenerife, it has previously been shown that road sides provide a suitable habitat for a hybrid swarm consisting of five different Argyranthemum species (Brochmann 1984; Francisco-Ortega et al. 2000). However, for Argyranthemum it is not clear whether the hybrids manage to expand from the road verges into the surrounding habitat (Francisco-Ortega et al. 2000). For Pericallis, such an expansion is obviously taking place since we found multiple admixed individuals outside the road verges.
Ongoing hybridization may lead to the permanent introgression of genes from one species into the other and may in extreme cases even result in a complete mixing of genomes (Levin et al. 1996; Rhymer and Simberloff 1996). Whether this will occur in Pericallis depends largely on whether the hybrid swarm will extend into the three core areas. Some of our results indicate that this is already happening: though the core areas still mostly consist of pure individuals, some of the individuals within the core areas show clear signs of admixture. This is for example the case for P. tussilaginis where there was one individual that showed both genetic and morphological signs of admixture. Another individual was genetically admixed but morphologically pure. In P. echinata, the patterns were more complex since there were multiple individuals with genetic signs of admixture even though they were morphologically indistinguishable from the other P. echinata individuals. Some of the mixed populations occur very close to the core areas. This is for example the case in populations 33 and 51, which are located in road verges near the hearts of the P. echinata and P. cruenta core areas, respectively. This further suggests that the roads can work as corridors that connect the core areas and promote hybridization. If the ongoing hybridization would result in a complete merging of the three Pericallis species, this would mean a further genetic erosion of the biodiversity of the Canary Islands (Francisco-Ortega et al. 2000).
The evolution of Pericallis in the context of the geological history of Tenerife
A phylogenetic study on Pericallis (Swenson and Manns 2003), based on ITS1 and ITS2 sequence data, showed the genus is of relatively recent evolutionary origin, but nevertheless shows a large morphological diversity. As with many adaptive radiations on oceanic islands (Okada et al. 1997; Böhle et al. 1996; Helfgott et al. 2000), it can be expected that the evolution of the genus is tightly linked with the geographic history of the archipelago. Until approximately 2 million years ago, Tenerife consisted of three separate volcanic islands, called Teno, Anaga and Roque del Conde. Between 1.9 and 0.2 Mya, these three islands were connected by lava flows emerging from the Cañadas volcano (the precursor of the present Mt. Teide) and represent the geologically oldest parts of Tenerife (Ancochea et al. 1990; Thirlwall et al. 2000). Because P. echinata has its core distribution area in Teno, whereas P. tussilaginis mainly occurs on Anaga, it is tempting to speculate that these species originated on these precursor islands. The P. cruenta core area is situated on much younger volcanic depositions in the region where around 0.8 Mya a chain of basaltic emission centers connected the Teide volcano with Anaga (Ancochea et al. 1990). This suggests that the colonization of Tenerife by this species has occurred relatively recently.
The Roque del Conde populations are located in the oldest region (11.6 Mya, Ancochea et al. 1990; Thirlwall et al. 2000) of the three precursor islands. Von Gaisberg and Stiersdorfer (2005) found that the highest numbers of endemics on the Canary island of El Hierro occur on such ancient geological outcrops. The outcrops that were not covered by more recent lava flows might have functioned as species refugia. The phylogeographic pattern of vicariant sister species affiliated with the Anaga and Teno massifs has also been reported for several animal species on Tenerife (reviewed in Juan et al. 2000). However, the unique morphology and genetic composition of the Roque del Conde populations might also be explained by founder events. The importance of such founder effects on the distribution of genetic variation in a Canarian endemic has been shown by García-Verdugo et al. (2010). Further molecular studies may shed light on the precise phylogenetic relations between the species, testing hypotheses that the three species either evolved in complete isolation, or that the Roque del Conde population is basal to all three other species.
Conclusions
We provide substantial morphological and genetic evidence for ongoing hybridization between P. cruenta and P. echinata as well as between P. cruenta and P. tussilaginis. The distribution of hybrid populations in road verges in between the parental core areas indicates that hybridization was promoted by secondary contact caused by human activities. This process may eventually threaten the existence of the three species and counteract the process of evolutionary radiation.
References
Ancochea E, Fuster JM, Ibarrola E, Cendrero A, Coello J, Hernan F, Cantagrel JM, Jamond C (1990) Volcanic evolution of the island of Tenerife (Canary-Islands) in the light of new K-Ar data. J Volcanol Geoth Res 44:231–249
Anderson E (1948) Hybridization of the habitat. Evolution 2:1–9
Baldwin BG, Crawford DJ, Francisco-Ortega J, Kin SC, Sang T, Stuessy TF (1998) Molecular phylogenetic insights on the origin and evolution of oceanic island plants. In: Soltis DE, Soltis P, Doyle JJ (eds) Molecular systematics of plants II. DNA sequencing. Kluwer, Boston, pp 410–441
Blair AC, Hufbauer RA (2010) Hybridization and invasion: one of North America’s most devastating invasive plants shows evidence for a history of interspecific hybridization. Evol Appl 3:40–51
Bleeker W, Hurka H (2001) Introgressive hybridization in Rorippa (Brassicaceae): gene flow and its consequences in natural and anthropogenic habitats. Mol Ecol 10:2013–2022
Böhle UR, Hilger HH, Martin WF (1996) Island colonization and evolution of the insular woody habit in Echium L. (Boraginaceae). P Natl Acad Sci USA 93:11740–11745
Bonin A, Belleman E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Bonin A, Ehrich D, Manel S (2007) Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16:3737–3758
Bramwell D, Bramwell Z (2001) Wild flowers of the Canary Islands. Aldington Books Ltd., Aldington
Brochmann C (1984) Hybridization and distribution of Argyranthemum coronopifolium (Asteraceae, Anthemideae) in the Canary Islands. Nord J Bot 4:729–736
Carlquist S, Cole MJ (1974) Island biology. Columbia University Press, New York
Caujapé-Castells J (2010) General G ST and θ inflation due to biased intra-population sampling, and its consequences for the conservation of the Canarian Flora. Conserv Genet 11:709–720
Delgado JD, Arroyo NL, Arévalo JR, Fernández-Palacios JM (2007) Edge effects of roads on temperature, light, canopy cover, and canopy height in laurel and pine forests (Tenerife, Canary Islands). Landscape Urban Plan 81:328–340
Dimitriadou E, Hornik K, Leisch F, Meyer D, Weingessel A (2010) e1071: Misc Functions of the Department of Statistics, TU Wien. R package version 1.5-24. http://CRAN.R-project.org/package=e1071
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–14
Dunn JC (1973) A fuzzy relative of the isodata process and its use in detecting compact well-separated clusters. J Cybernetics 3:32–57
Emerson BC (2002) Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol Ecol 11:951–966
Esa YB, Waters JM, Wallis GP (2000) Introgressive hybridization between Galaxias depressiceps and Galaxias sp D (Teleostei: Galaxiidae) in Otago, New Zealand: secondary contact mediated by water races. Conserv Genet 1:329–339
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 4:574–578
Fjellheim S, Jorgensen MH, Kjos M, Borgen L (2009) A molecular study of hybridization and homoploid hybrid speciation in Argyranthemum (Asteraceae) on Tenerife, the Canary Islands. Bot J Linn Soc 159:19–31
Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ (2000) Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87:909–919
François O, Ancelet S, Guillot G (2006) Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 174:805–816
García-Verdugo C, Forrest AD, Fay MF, Vargas P (2010) The relevance of gene flow in metapopulation dynamics of an oceanic island endemic Olea europaea subsp. guanchica. Evolution 64–12:3525–3536
Gobierno de Canarias. Banco de Datos de Biodiversidad de Canarias (http://www.biodiversidadcanarias.es). Consulted in 2011
Gower JC (1966) Some distant properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electronica 4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.html
Helfgott DM, Francisco-Ortega J, Santos-Guerra A, Jansen RK, Simpson BB (2000) Biogeography and breeding system evolution of the woody Bencomia alliance (Rosaceae) in Macaronesia based on ITS sequence data. Syst Bot 25:82–97
Hodkinson DJ, Thompson K (1997) Plant dispersal: the role of man. J Appl Ecol 34:1484–1496
Hohenester A, Welss W (1993) Exkursionsflora für die Kanarischen Inseln: mit Ausblicken auf ganz Makaronesien. Eugen Ulmer Verlag, Stuttgart
Juan C, Emerson BC, Oromi P, Hewitt GM (2000) Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends EcolEvol 15:104–109
Kalinowski S (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106(4):625–632
Kornerup A, Wanscher JH (1967) Methuen handbook of colour. Methuen, London
Lamont BB, He T, Enright NJ, Krauss SL, Miller BP (2003) Anthropogenic disturbance promotes hybridization between Banksia species by altering their biology. J Evolution Biol 16:551–557
Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto A, Kremer A, Gerber S (2009) Species relative abundance and direction of introgression in oaks. Mol Ecol 18:2228–2242
Levin DA, Francisco-Ortega J, Jansen RK (1996) Hybridization and the extinction of rare plant species. Conserv Biol 10:10–16
Meirmans PG, Hedrick PW (2011) Assessing population structure: FST and related measures. Mol Ecol Resour 11:5–18
Meirmans PG, Van Tienderen PH (2004) Genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Mráz P, Chrtek J, Fehrer J (2011) Interspecific hybridization in the genus Hieracium s. str.: evidence for bidirectional gene flow and spontaneous allopolyploidization. Plant Syst Evol 293:237–245
Nordenstam B (1978) Taxonomic studies in the tribe Senecioneae (Compositae). Opera Bot 44:1–84
Okada M, Whitkus R, Lowrey TK (1997) Genetics of adaptive radiation in Hawaiian and Cook Islands species of Tetramolopium (Asteraceae; Astereae) 1. Nuclear RFLP marker diversity. Am J Bot 84:1236–1246
Oostermeijer JGB, Luijten SH, den Nijs JCM (2003) Integrating demographic and genetic approaches in plant conservation. Biol Conserv 113:389–398
Panero JL, Francisco-Ortega J, Jansen RK, Santos-Guerra A (1999) Molecular evidence for multiple origins of woodiness and a new world biogeographic connection of the Macaronesian Island endemic Pericallis (Asteraceae : Senecioneae). P Natl Acad Sci USA 96:13886–13891
Pott R, Hüppe J, Wildpret de la Torre W (2003) Die Kanarischen Inseln. Eugen Ulmer Verlag, Stuttgart
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0, URL http://www.R-project.org
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109
Rivas-Martínez S, Wildpret de la Torre W, Del Arco AguilarM, Rodríguez O, Pérez de Paz PL, Garcia Gallo A, Acebes Ginovés JR et al (1993) Las comunidades vegetales de la Isla de Tenerife (Islas Canarias). Itinera Geobotanica 7:169–374
Schierenbeck KA, Ellstrand NC (2009) Hybridization and the evolution of invasiveness in plants and other organisms. Biol Invasions 11:1093–1105
Schluter D (2000) The ecology of adaptive radiation. Oxford University Press Inc., New York
Schönfelder P, Schönfelder I (1997) Die Kosmos-Kanarenflora: über 850 Arten der Kanarenflora. Kosmos, Stuttgart
Schultz CH (1845–1846) Doronicum. Linn. ex parte. In: Webb PB, Berthelot S (eds) Histoire naturelle des îles Canaries, 3(2): Botanique. Phytogeographia Canariensis, Paris, pp 330–340
Suda J, Kyncl T, Freiová R (2003) Nuclear DNA amounts in Macaronesian angiosperms. Ann Bot-London 92:153–164
Suda J, Kyncl T, Jarolímová V (2005) Genome size variation in Macaronesian angiosperms: forty percent of the Canarian endemic flora completed. Plant Syst Evol 252:215–238
Swenson U, Manns U (2003) Phylogeny of Pericallis (Asteraceae): a total evidence approach reappraising the double origin of woodiness. Taxon 52:533–546
Thirlwall MF, Singer BS, Marriner GF (2000) Ar-39-Ar-40 ages and geochemistry of the basaltic shield stage of Tenerife, Canary Islands, Spain. J Volcanol Geoth Res 103:247–297
Thompson S, Lamothe M, Meirmans PG, Perinet P, Isabel N (2010) Repeated unidirectional introgression towards Populus balsamifera in contact zones of exotic and native poplars. Mol Ecol 19:132–145
Urbanska KM, Hurka H, Landolt E, Neuffer B, Mummenhoff K (1997) Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, Central Switzerland: biosystematic and molecular evidence. Plant Syst Evol 204:233–256
Vaha JP, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15:63–72
Von der Lippe M, Kowarik I (2007) Long-distance dispersal of plants by vehicles as a driver of plant invasions. Conserv Biol 21:986–996
Von Gaisberg M, Stierstorfer C (2005) The significance of geological traits for the speciation of endemic angiosperms on El Hierro (Canary Islands). Phytocoenologia 35:39–52
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A et al (1995) AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res 23:4407–4414
Whittaker RJ, Fernández-Palacios JM (2007) Island Biogeography: ecology, evolution, and conservation, 2nd edn. Oxford University Press, New York
Wichmann MC, Alexander MJ, Soons MB, Galsworthy S, Dunne L, Gould R, Fairfax C et al (2009) Human-mediated dispersal of seeds over long distances. P Roy Soc B-Biol Sci 276:523–532
Young AG, Clarke GM (2000) Genetics, demography and viability of fragmented populations. Cambridge University Press, Cambridge
Acknowledgments
The authors thank the students of the course Evolutionary Botany (University of Amsterdam, IBED) for their help in collecting and analyzing many of the samples. Prof. Dr. M. Del Arco Aguilar and Dr. J.R. Acebes Ginovés (Universidad de La Laguna) gave highly appreciated advice as well as access to the La Laguna Herbarium. Their help during collecting trips is also very much acknowledged. Thanks are also due to P. Kuperus for his essential assistance in the laboratory and R. Bregman for performing interspecific crosses in the greenhouse.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
van Hengstum, T., Lachmuth, S., Oostermeijer, J.G.B. et al. Human-induced hybridization among congeneric endemic plants on Tenerife, Canary Islands. Plant Syst Evol 298, 1119–1131 (2012). https://doi.org/10.1007/s00606-012-0624-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0624-6